Gwaihir: Jupyter Notebook graphical user interface for Bragg coherent diffraction imaging
- PMID: 35974722
- PMCID: PMC9348885
- DOI: 10.1107/S1600576722005854
Gwaihir: Jupyter Notebook graphical user interface for Bragg coherent diffraction imaging
Abstract
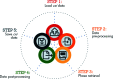
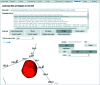
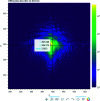
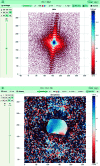
Bragg coherent X-ray diffraction is a nondestructive method for probing material structure in three dimensions at the nanoscale, with unprecedented resolution in displacement and strain fields. This work presents Gwaihir, a user-friendly and open-source tool to process and analyze Bragg coherent X-ray diffraction data. It integrates the functionalities of the existing packages bcdi and PyNX in the same toolbox, creating a natural workflow and promoting data reproducibility. Its graphical interface, based on Jupyter Notebook widgets, combines an interactive approach for data analysis with a powerful environment designed to link large-scale facilities and scientists.
Keywords: Jupyter Notebook; X-ray diffraction; coherence; graphical user interfaces; phase retrieval.
© David Simonne et al. 2022.
Figures






References
-
- Ahrens, J., Geveci, B. & Law, C. (2005). ParaView: An End-User Tool for Large Data Visualization, The Visualization Handbook, Vol. 717. Munich: Elsevier.
-
- Baker, M. (2016). Nature, 533, 452–454. - PubMed
-
- Bokeh Development Team (2018). Bokeh, https://bokeh.pydata.org/en/latest/.
-
- Boulle, A. & Kieffer, J. (2019). J. Appl. Cryst. 52, 882–897.
-
- Breddeld, M. (2021). ipyvolume, version 0.6.0, https://github.com/maartenbreddels/ipyvolume.
LinkOut - more resources
Full Text Sources
