Labeling and sequencing nucleic acid modifications using bio-orthogonal tools
- PMID: 35975003
- PMCID: PMC9347354
- DOI: 10.1039/d2cb00087c
Labeling and sequencing nucleic acid modifications using bio-orthogonal tools
Abstract
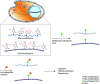
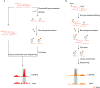
The bio-orthogonal reaction is a type of reaction that can occur within a cell without interfering with the active components of the cell. Bio-orthogonal reaction techniques have been used to label and track the synthesis, metabolism, and interactions of distinct biomacromolecules in cells. Thus, it is a handy tool for analyzing biological macromolecules within cells. Nucleic acid modifications are widely distributed in DNA and RNA in cells and play a critical role in regulating physiological and pathological cellular activities. Utilizing bio-orthogonal tools to study modified bases is a critical and worthwhile research direction. The development of bio-orthogonal reactions focusing on nucleic acid modifications has enabled the mapping of nucleic acid modifications in DNA and RNA. This review discusses the recent advances in bio-orthogonal labeling and sequencing nucleic acid modifications in DNA and RNA.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures









Similar articles
-
Bio-Orthogonal Mediated Nucleic Acid Transfection of Cells via Cell Surface Engineering.ACS Cent Sci. 2017 May 24;3(5):489-500. doi: 10.1021/acscentsci.7b00132. Epub 2017 May 15. ACS Cent Sci. 2017. PMID: 28573212 Free PMC article.
-
Detection of nucleic acid modifications by chemical reagents.RNA Biol. 2017 Sep 2;14(9):1166-1174. doi: 10.1080/15476286.2016.1261788. Epub 2016 Nov 30. RNA Biol. 2017. PMID: 27901634 Free PMC article. Review.
-
Recent Developments in Metal-Catalyzed Bio-orthogonal Reactions for Biomolecule Tagging.Chembiochem. 2019 Jun 14;20(12):1498-1507. doi: 10.1002/cbic.201900052. Epub 2019 Apr 15. Chembiochem. 2019. PMID: 30676678 Review.
-
Dynamic modifications of biomacromolecules: mechanism and chemical interventions.Sci China Life Sci. 2019 Nov;62(11):1459-1471. doi: 10.1007/s11427-019-9823-1. Epub 2019 Sep 18. Sci China Life Sci. 2019. PMID: 31555961 Review.
-
Design and Engineering of Metal Catalysts for Bio-orthogonal Catalysis in Living Systems.ACS Appl Bio Mater. 2020 Aug 17;3(8):4717-4746. doi: 10.1021/acsabm.0c00581. Epub 2020 Jul 27. ACS Appl Bio Mater. 2020. PMID: 35021720
Cited by
-
Qualitative and Quantitative Analytical Techniques of Nucleic Acid Modification Based on Mass Spectrometry for Biomarker Discovery.Int J Mol Sci. 2024 Mar 16;25(6):3383. doi: 10.3390/ijms25063383. Int J Mol Sci. 2024. PMID: 38542356 Free PMC article. Review.
-
Not So Bioorthogonal Chemistry.J Am Chem Soc. 2025 Mar 12;147(10):8049-8062. doi: 10.1021/jacs.4c15986. Epub 2025 Feb 28. J Am Chem Soc. 2025. PMID: 40017419 Free PMC article. Review.
-
Past, Present, and Future of RNA Modifications in Infectious Disease Research.ACS Infect Dis. 2024 Dec 13;10(12):4017-4029. doi: 10.1021/acsinfecdis.4c00598. Epub 2024 Nov 21. ACS Infect Dis. 2024. PMID: 39569943
-
Unprecedented reactivity of polyamines with aldehydic DNA modifications: structural determinants of reactivity, characterization and enzymatic stability of adducts.Nucleic Acids Res. 2023 Nov 10;51(20):10846-10866. doi: 10.1093/nar/gkad837. Nucleic Acids Res. 2023. PMID: 37850658 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

