Quantitative, multiplexed, targeted proteomics for ascertaining variant specific SARS-CoV-2 antibody response
- PMID: 35975199
- PMCID: PMC9372021
- DOI: 10.1016/j.crmeth.2022.100279
Quantitative, multiplexed, targeted proteomics for ascertaining variant specific SARS-CoV-2 antibody response
Abstract
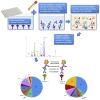
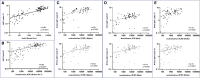
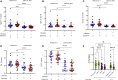
Determining the protection an individual has to severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) variants of concern (VoCs) is crucial for future immune surveillance, vaccine development, and understanding of the changing immune response. We devised an informative assay to current ELISA-based serology using multiplexed, baited, targeted proteomics for direct detection of multiple proteins in the SARS-CoV-2 anti-spike antibody immunocomplex. Serum from individuals collected after infection or first- and second-dose vaccination demonstrates this approach and shows concordance with existing serology and neutralization. Our assays show altered responses of both immunoglobulins and complement to the Alpha (B.1.1.7), Beta (B.1.351), and Delta (B.1.617.1) VoCs and a reduced response to Omicron (B1.1.1529). We were able to identify individuals who had prior infection, and observed that C1q is closely associated with IgG1 (r > 0.82) and may better reflect neutralization to VoCs. Analyzing additional immunoproteins beyond immunoglobulin (Ig) G, provides important information about our understanding of the response to infection and vaccination.
Trial registration: ClinicalTrials.gov NCT04318314.
Keywords: COVID-19; SARS-CoV-2; complement: immunoglobulin; delta variant; mass spectrometry; omicron variant; proteomics; vaccination; variant of concern.
© 2022 The Author(s).
Conflict of interest statement
The authors have submitted an intellectual property claim for using the technology for clinical applications.
Figures



References
-
- Abbasi J. The flawed science of antibody testing for SARS-CoV-2 immunity. JAMA. 2021;326:1781–1782. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

