Pigmentation biosynthesis influences the microbiome in sea urchins
- PMID: 35975446
- PMCID: PMC9382222
- DOI: 10.1098/rspb.2022.1088
Pigmentation biosynthesis influences the microbiome in sea urchins
Abstract
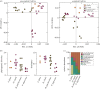
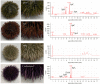
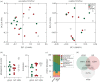
Organisms living on the seafloor are subject to encrustations by a wide variety of animals, plants and microbes. Sea urchins, however, thwart this covering. Despite having a sophisticated immune system, there is no clear molecular mechanism that allows sea urchins to remain free of epibiotic microorganisms. Here, we test the hypothesis that pigmentation biosynthesis in sea urchin spines influences their interactions with microbes in vivo using CRISPR/Cas9. We report three primary findings. First, the microbiome of sea urchin spines is species-specific and much of this community is lost in captivity. Second, different colour morphs associate with bacterial communities that are similar in taxonomic composition, diversity and evenness. Lastly, loss of the pigmentation biosynthesis genes polyketide synthase and flavin-dependent monooxygenase induces a shift in which bacterial taxa colonize sea urchin spines. Therefore, our results are consistent with the hypothesis that host pigmentation biosynthesis can, but may not always, influence the microbiome in sea urchin spines.
Keywords: echinoderm; flavin-dependent monooxygenase; host–microbe symbiosis; microbial communities; polyketide pigments; polyketide synthase.
Conflict of interest statement
We declare we have no competing interests.
Figures


References
-
- Hyman L. 1955. The invertebrates. New York, NY: McGraw-Hill.
-
- Smith L, et al. 2018. Echinodermata: the complex immune system in echinoderms. In Advances in comparative immunology (ed. Cooper E), pp. 409-501. Berlin, Germany: Springer.
-
- O'Hara T, Byrne M. 2017. Australian echinoderms: biology, ecology and evolution. Victoria, Australia: CSIRO Publishing.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous
