Bacterial degrons in synthetic circuits
- PMID: 35975648
- PMCID: PMC9382460
- DOI: 10.1098/rsob.220180
Bacterial degrons in synthetic circuits
Abstract
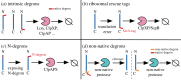
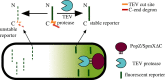
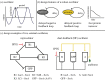
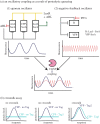
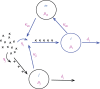
Bacterial proteases are a promising post-translational regulation strategy in synthetic circuits because they recognize specific amino acid degradation tags (degrons) that can be fine-tuned to modulate the degradation levels of tagged proteins. For this reason, recent efforts have been made in the search for new degrons. Here we review the up-to-date applications of degradation tags for circuit engineering in bacteria. In particular, we pay special attention to the effects of degradation bottlenecks in synthetic oscillators and introduce mathematical approaches to study queueing that enable the quantitative modelling of proteolytic queues.
Keywords: degradation; oscillatory circuits; proteases; queueing theory.
Conflict of interest statement
We declare we have no competing interests.
Figures


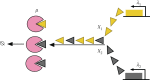
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
