Controlling amyloid formation of intrinsically disordered proteins and peptides: slowing down or speeding up?
- PMID: 35975807
- PMCID: PMC7617668
- DOI: 10.1042/EBC20220046
Controlling amyloid formation of intrinsically disordered proteins and peptides: slowing down or speeding up?
Abstract
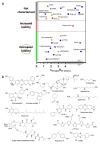
The pathological assembly of intrinsically disordered proteins/peptides (IDPs) into amyloid fibrils is associated with a range of human pathologies, including neurodegeneration, metabolic diseases and systemic amyloidosis. These debilitating disorders affect hundreds of millions of people worldwide, and the number of people affected is increasing sharply. However, the discovery of therapeutic agents has been immensely challenging largely because of (i) the diverse number of aggregation pathways and the multi-conformational and transient nature of the related proteins or peptides and (ii) the under-development of experimental pipelines for the identification of disease-modifying molecules and their mode-of-action. Here, we describe current approaches used in the search for small-molecule modulators able to control or arrest amyloid formation commencing from IDPs and review recently reported accelerators and inhibitors of amyloid formation for this class of proteins. We compare their targets, mode-of-action and effects on amyloid-associated cytotoxicity. Recent successes in the control of IDP-associated amyloid formation using small molecules highlight exciting possibilities for future intervention in protein-misfolding diseases, despite the challenges of targeting these highly dynamic precursors of amyloid assembly.
Keywords: accelerator; amyloid; energy landscape; inhibitor; protein aggregation; small-molecule modulator.
© 2022 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
All authors declare they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

