Insights from the genomes of 4 diploid Camelina spp
- PMID: 35976116
- PMCID: PMC9713399
- DOI: 10.1093/g3journal/jkac182
Insights from the genomes of 4 diploid Camelina spp
Abstract
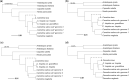
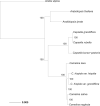
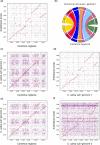
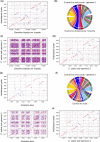
Plant evolution has been a complex process involving hybridization and polyploidization making understanding the origin and evolution of a plant's genome challenging even once a published genome is available. The oilseed crop, Camelina sativa (Brassicaceae), has a fully sequenced allohexaploid genome with 3 unknown ancestors. To better understand which extant species best represent the ancestral genomes that contributed to C. sativa's formation, we sequenced and assembled chromosome level draft genomes for 4 diploid members of Camelina: C. neglecta C. hispida var. hispida, C. hispida var. grandiflora, and C. laxa using long and short read data scaffolded with proximity data. We then conducted phylogenetic analyses on regions of synteny and on genes described for Arabidopsis thaliana, from across each nuclear genome and the chloroplasts to examine evolutionary relationships within Camelina and Camelineae. We conclude that C. neglecta is closely related to C. sativa's sub-genome 1 and that C. hispida var. hispida and C. hispida var. grandiflora are most closely related to C. sativa's sub-genome 3. Further, the abundance and density of transposable elements, specifically Helitrons, suggest that the progenitor genome that contributed C. sativa's sub-genome 3 maybe more similar to the genome of C. hispida var. hispida than that of C. hispida var. grandiflora. These diploid genomes show few structural differences when compared to C. sativa's genome indicating little change to chromosome structure following allopolyploidization. This work also indicates that C. neglecta and C. hispida are important resources for understanding the genetics of C. sativa and potential resources for crop improvement.
Keywords: Brassicaceae; Camelina; allopolyploidy; chromosome evolution; genome evolution; phylogenomics.
© Her Majesty the Queen in Right of Canada 2022, as represented by the Minister of Agriculture and Agri-Food Canada.
Figures




Similar articles
-
Sequencing of Camelina neglecta, a diploid progenitor of the hexaploid oilseed Camelina sativa.Plant Biotechnol J. 2023 Mar;21(3):521-535. doi: 10.1111/pbi.13968. Epub 2022 Dec 12. Plant Biotechnol J. 2023. PMID: 36398722 Free PMC article.
-
Origin and Evolution of Diploid and Allopolyploid Camelina Genomes Were Accompanied by Chromosome Shattering.Plant Cell. 2019 Nov;31(11):2596-2612. doi: 10.1105/tpc.19.00366. Epub 2019 Aug 26. Plant Cell. 2019. PMID: 31451448 Free PMC article.
-
The identification of the missing maternal genome of the allohexaploid camelina (Camelina sativa).Plant J. 2022 Nov;112(3):622-629. doi: 10.1111/tpj.15931. Epub 2022 Oct 4. Plant J. 2022. PMID: 35916590
-
Camelina sativa, an oilseed at the nexus between model system and commercial crop.Plant Cell Rep. 2018 Oct;37(10):1367-1381. doi: 10.1007/s00299-018-2308-3. Epub 2018 Jun 7. Plant Cell Rep. 2018. PMID: 29881973 Review.
-
Overcoming genetic paucity of Camelina sativa: possibilities for interspecific hybridization conditioned by the genus evolution pathway.Front Plant Sci. 2023 Sep 25;14:1259431. doi: 10.3389/fpls.2023.1259431. eCollection 2023. Front Plant Sci. 2023. PMID: 37818316 Free PMC article. Review.
Cited by
-
Chromosome-level assembly and analysis of Camelina neglecta: a novel diploid model for Camelina biotechnology research.Biotechnol Biofuels Bioprod. 2024 Jan 31;17(1):17. doi: 10.1186/s13068-024-02466-9. Biotechnol Biofuels Bioprod. 2024. PMID: 38291537 Free PMC article.
-
Exploring genetic diversity, population structure, and subgenome differences in the allopolyploid Camelina sativa: implications for future breeding and research studies.Hortic Res. 2024 Sep 9;11(11):uhae247. doi: 10.1093/hr/uhae247. eCollection 2024 Nov. Hortic Res. 2024. PMID: 39539416 Free PMC article.
-
Sequencing of Camelina neglecta, a diploid progenitor of the hexaploid oilseed Camelina sativa.Plant Biotechnol J. 2023 Mar;21(3):521-535. doi: 10.1111/pbi.13968. Epub 2022 Dec 12. Plant Biotechnol J. 2023. PMID: 36398722 Free PMC article.
-
Genome-wide identification and diversity of FAD2, FAD3 and FAE1 genes in terms of biotechnological importance in Camelina species.BMC Biotechnol. 2024 Dec 18;24(1):107. doi: 10.1186/s12896-024-00936-4. BMC Biotechnol. 2024. PMID: 39695603 Free PMC article.
-
Chromosome fusions shaped karyotype evolution and evolutionary relationships in the model family Brassicaceae.Nat Commun. 2025 May 19;16(1):4631. doi: 10.1038/s41467-025-59640-2. Nat Commun. 2025. PMID: 40389407 Free PMC article.
References
-
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, et al.Hybridization and speciation. J Evol Biol. 2013;26(2):229–246. doi:10.1111/j.1420–9101.2012.02599.x. - PubMed
-
- Al-Shehbaz IA. A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon. 2012;61(5):931–954.
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous
