A continental-scale survey of Wolbachia infections in blue butterflies reveals evidence of interspecific transfer and invasion dynamics
- PMID: 35976120
- PMCID: PMC9526071
- DOI: 10.1093/g3journal/jkac213
A continental-scale survey of Wolbachia infections in blue butterflies reveals evidence of interspecific transfer and invasion dynamics
Abstract
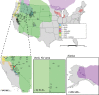
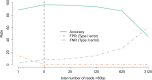
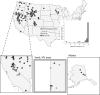
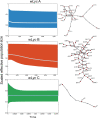
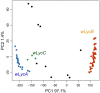
Infections by maternally inherited bacterial endosymbionts, especially Wolbachia, are common in insects and other invertebrates but infection dynamics across species ranges are largely under studied. Specifically, we lack a broad understanding of the origin of Wolbachia infections in novel hosts, and the historical and geographical dynamics of infections that are critical for identifying the factors governing their spread. We used Genotype-by-Sequencing data from previous population genomics studies for range-wide surveys of Wolbachia presence and genetic diversity in North American butterflies of the genus Lycaeides. As few as one sequence read identified by assembly to a Wolbachia reference genome provided high accuracy in detecting infections in host butterflies as determined by confirmatory PCR tests, and maximum accuracy was achieved with a threshold of only 5 sequence reads per host individual. Using this threshold, we detected Wolbachia in all but 2 of the 107 sampling localities spanning the continent, with infection frequencies within populations ranging from 0% to 100% of individuals, but with most localities having high infection frequencies (mean = 91% infection rate). Three major lineages of Wolbachia were identified as separate strains that appear to represent 3 separate invasions of Lycaeides butterflies by Wolbachia. Overall, we found extensive evidence for acquisition of Wolbachia through interspecific transfer between host lineages. Strain wLycC was confined to a single butterfly taxon, hybrid lineages derived from it, and closely adjacent populations in other taxa. While the other 2 strains were detected throughout the rest of the continent, strain wLycB almost always co-occurred with wLycA. Our demographic modeling suggests wLycB is a recent invasion. Within strain wLycA, the 2 most frequent haplotypes are confined almost exclusively to separate butterfly taxa with haplotype A1 observed largely in Lycaeides melissa and haplotype A2 observed most often in Lycaeides idas localities, consistent with either cladogenic mode of infection acquisition from a common ancestor or by hybridization and accompanying mutation. More than 1 major Wolbachia strain was observed in 15 localities. These results demonstrate the utility of using resequencing data from hosts to quantify Wolbachia genetic variation and infection frequency and provide evidence of multiple colonizations of novel hosts through hybridization between butterfly lineages and complex dynamics between Wolbachia strains.
Keywords: Lycaeides; Wolbachia; GBS data; geography of infection; host parasite interactions; infection acquisition.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures

Similar articles
-
Widespread mito-nuclear discordance with evidence for introgressive hybridization and selective sweeps in Lycaeides.Mol Ecol. 2008 Dec;17(24):5231-44. doi: 10.1111/j.1365-294X.2008.03988.x. Mol Ecol. 2008. PMID: 19120997
-
Genetic structure of sibling butterfly species affected by Wolbachia infection sweep: evolutionary and biogeographical implications.Mol Ecol. 2006 Apr;15(4):1095-108. doi: 10.1111/j.1365-294X.2006.02857.x. Mol Ecol. 2006. PMID: 16599969
-
The joint evolutionary histories of Wolbachia and mitochondria in Hypolimnas bolina.BMC Evol Biol. 2009 Mar 24;9:64. doi: 10.1186/1471-2148-9-64. BMC Evol Biol. 2009. PMID: 19317891 Free PMC article.
-
Distribution and evolutionary impact of wolbachia on butterfly hosts.Indian J Microbiol. 2014 Sep;54(3):249-54. doi: 10.1007/s12088-014-0448-x. Epub 2014 Feb 9. Indian J Microbiol. 2014. PMID: 24891730 Free PMC article. Review.
-
Wolbachia strains for disease control: ecological and evolutionary considerations.Evol Appl. 2015 Sep;8(8):751-68. doi: 10.1111/eva.12286. Epub 2015 Jul 20. Evol Appl. 2015. PMID: 26366194 Free PMC article. Review.
Cited by
-
Global determinants of insect mitochondrial genetic diversity.Nat Commun. 2023 Aug 29;14(1):5276. doi: 10.1038/s41467-023-40936-0. Nat Commun. 2023. PMID: 37644003 Free PMC article.
-
Assessing Wolbachia circulation in wild populations of phlebotomine sand flies from Spain and Morocco: implications for control of leishmaniasis.Parasit Vectors. 2025 Apr 26;18(1):155. doi: 10.1186/s13071-025-06771-6. Parasit Vectors. 2025. PMID: 40287743 Free PMC article.
-
Pervasive horizontal transmission of Wolbachia in natural populations of closely related and widespread tropical skipper butterflies.BMC Microbiol. 2025 Jan 7;25(1):5. doi: 10.1186/s12866-024-03719-1. BMC Microbiol. 2025. PMID: 39773184 Free PMC article.
References
-
- Aktas C. haplotypes: Manipulating DNA Sequences and Estimating Unambiguous Haplotype Network with Statistical Parsimony. R Package Version 1.1.2; 2020. (https://cran.r-project.org/web/packages/haplotypes/index.html)
-
- Arif S, Gerth M, Hone-Millard WG, Nunes MD, Dapporto L, Shreeve TG.. Evidence for multiple colonisations and Wolbachia infections shaping the genetic structure of the widespread butterfly Polyommatus icarus in the British Isles. Mol Ecol. 2021;30(20):5196–5213. - PubMed
-
- Bailly-Bechet M, Martins-Simões P, Szöllosi GJ, Mialdea G, Sagot M-F, Charlat S.. How long does Wolbachia remain on board? Mol Biol Evol. 2017;34(5):1183–1193. - PubMed
-
- Baldo L, Ayoub NA, Hayashi CY, Russell JA, Stahlhut JK, Werren JH.. Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol Ecol. 2008;17(2):557–569. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
