The renaissance and enlightenment of Marchantia as a model system
- PMID: 35976122
- PMCID: PMC9516144
- DOI: 10.1093/plcell/koac219
The renaissance and enlightenment of Marchantia as a model system
Abstract
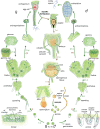
The liverwort Marchantia polymorpha has been utilized as a model for biological studies since the 18th century. In the past few decades, there has been a Renaissance in its utilization in genomic and genetic approaches to investigating physiological, developmental, and evolutionary aspects of land plant biology. The reasons for its adoption are similar to those of other genetic models, e.g. simple cultivation, ready access via its worldwide distribution, ease of crossing, facile genetics, and more recently, efficient transformation, genome editing, and genomic resources. The haploid gametophyte dominant life cycle of M. polymorpha is conducive to forward genetic approaches. The lack of ancient whole-genome duplications within liverworts facilitates reverse genetic approaches, and possibly related to this genomic stability, liverworts possess sex chromosomes that evolved in the ancestral liverwort. As a representative of one of the three bryophyte lineages, its phylogenetic position allows comparative approaches to provide insights into ancestral land plants. Given the karyotype and genome stability within liverworts, the resources developed for M. polymorpha have facilitated the development of related species as models for biological processes lacking in M. polymorpha.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures


References
-
- Aguilar-cruz A, Grimanelli D, Haseloff J, Arteaga-vazquez MA (2019) DNA methylation in Marchantia polymorpha. New Phytol 223: 575–581 - PubMed
-
- Aki SS, Mikami T, Naramoto S, Nishihama R, Ishizaki K, Kojima M, Takebayashi Y, Sakakibara H, Kyozuka J, Kohchi T, et al. (2019) Cytokinin signaling is essential for organ formation in Marchantia polymorpha. Plant & Cell Physiol 60: 1842–1854 - PubMed
-
- Albert NW, Thrimawithana AH, Mcghie TK, Clayton WA, Deroles SC, Schwinn KE, Bowman JL, Jordan BR, Davies KM (2018) Genetic analysis of the liverwort Marchantia polymorpha reveals that R2R3MYB activation of flavonoid production in response to abiotic stress is an ancient character in land plants. New Phytol 218: 554–566 - PubMed
-
- Allen CE (1917) A chromosome difference correlated with sex differences in Sphærocarpos. Science 46: 466–467 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

