The photosynthesis apparatus of European mistletoe (Viscum album)
- PMID: 35976139
- PMCID: PMC9614478
- DOI: 10.1093/plphys/kiac377
The photosynthesis apparatus of European mistletoe (Viscum album)
Abstract
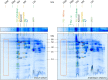
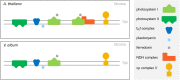
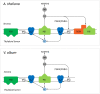
European mistletoe (Viscum album) is known for its special mode of cellular respiration. It lacks the mitochondrial NADH dehydrogenase complex (Complex I of the respiratory chain) and has restricted capacities to generate mitochondrial adenosine triphosphate (ATP). Here, we present an investigation of the V. album energy metabolism taking place in chloroplasts. Thylakoids were purified from young V. album leaves, and membrane-bound protein complexes were characterized by Blue native polyacrylamide gel electrophoresis as well as by the complexome profiling approach. Proteins were systematically identified by label-free quantitative shotgun proteomics. We identified >1,800 distinct proteins (accessible at https://complexomemap.de/va_leaves), including nearly 100 proteins forming part of the protein complexes involved in the light-dependent part of photosynthesis. The photosynthesis apparatus of V. album has distinct features: (1) comparatively low amounts of Photosystem I; (2) absence of the NDH complex (the chloroplast pendant of mitochondrial Complex I involved in cyclic electron transport (CET) around Photosystem I); (3) reduced levels of the proton gradient regulation 5 (PGR5) and proton gradient regulation 5-like 1 (PGRL1) proteins, which offer an alternative route for CET around Photosystem I; (4) comparable amounts of Photosystem II and the chloroplast ATP synthase complex to other seed plants. Our data suggest a restricted capacity for chloroplast ATP biosynthesis by the photophosphorylation process. This is in addition to the limited ATP supply by the mitochondria. We propose a view on mistletoe's mode of life, according to which its metabolism relies to a greater extent on energy-rich compounds provided by the host trees.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures







References
-
- Arike L, Valgepea K, Peil L, Nahku R, Adamberg K, Vilu R (2012) Comparison and applications of label-free absolute proteome quantification methods on Escherichia coli. J Proteomics 75: 5437–5448 - PubMed
-
- Arnold S, Braun HP (2022) The complexome profiling approach for direct biochemical analysis of multiprotein assemblies. Biochim Biophys Acta Bioenerg 1863: 148522. - PubMed
-
- Becker H (1986) Botany of European mistletoe (Viscum album L.). Oncology 43 Suppl 1: 2–7 - PubMed
-
- Behrens C, Blume C, Senkler M, Eubel H, Peterhänsel C, Braun HP (2013) The ‘protein complex proteome’ of chloroplasts in Arabidopsis thaliana. J Proteomics 91: 73–83 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

