A retrotransposon insertion in MUTL-HOMOLOG 1 affects wild rice seed set and cultivated rice crossover rate
- PMID: 35976143
- PMCID: PMC9614510
- DOI: 10.1093/plphys/kiac378
A retrotransposon insertion in MUTL-HOMOLOG 1 affects wild rice seed set and cultivated rice crossover rate
Abstract
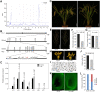
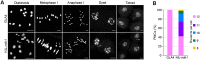
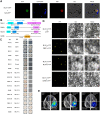
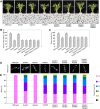
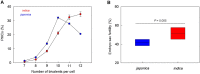
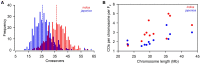
Wild rice (Oryza rufipogon) has a lower panicle seed setting rate (PSSR) and gamete fertility than domesticated rice (Oryza sativa), but the genetic mechanisms of this phenomenon remain unknown. Here, we cloned a null allele of OsMLH1, an ortholog of MutL-homolog 1 to yeast and mammals, from wild rice O. rufipogon W1943 and revealed a 5.4-kb retrotransposon insertion in OsMLH1 is responsible for the low PSSR in wild rice. In contrast to the wild-type, a near isogenic line NIL-mlh1 exhibits defective crossover (CO) formation during meiosis, resulting in reduced pollen viability, partial embryo lethality, and low PSSR. Except for the mutant of mismatch repair gene postmeiotic segregation 1 (Ospms1), all other MutL mutants from O. sativa indica subspecies displayed male and female semi-sterility similar to NIL-mlh1, but less severe than those from O. sativa japonica subspecies. MLH1 and MLH3 did not contribute in an additive fashion to fertility. Two types of MutL heterodimers, MLH1-PMS1 and MLH1-MLH3, were identified in rice, but only the latter functions in promoting meiotic CO formation. Compared to japonica varieties, indica cultivars had greater numbers of CO events per meiosis. Our results suggest that low fertility in wild rice may be caused by different gene defects, and indica and japonica subspecies have substantially different CO rates responsible for the discrepancy between the fertility of mlh1 and mlh3 mutants.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


Similar articles
-
Rice MutLγ, the MLH1-MLH3 heterodimer, participates in the formation of type I crossovers and regulation of embryo sac fertility.Plant Biotechnol J. 2021 Jul;19(7):1443-1455. doi: 10.1111/pbi.13563. Epub 2021 Feb 22. Plant Biotechnol J. 2021. PMID: 33544956 Free PMC article.
-
OsMLH1 interacts with OsMLH3 to regulate synapsis and interference-sensitive crossover formation during meiosis in rice.J Genet Genomics. 2021 Jun 20;48(6):485-496. doi: 10.1016/j.jgg.2021.04.011. Epub 2021 Jun 7. J Genet Genomics. 2021. PMID: 34257043
-
mlh3 mutations in baker's yeast alter meiotic recombination outcomes by increasing noncrossover events genome-wide.PLoS Genet. 2017 Aug 21;13(8):e1006974. doi: 10.1371/journal.pgen.1006974. eCollection 2017 Aug. PLoS Genet. 2017. PMID: 28827832 Free PMC article.
-
Expanded roles for the MutL family of DNA mismatch repair proteins.Yeast. 2021 Jan;38(1):39-53. doi: 10.1002/yea.3512. Epub 2020 Jul 30. Yeast. 2021. PMID: 32652606 Free PMC article. Review.
-
Coordinated and Independent Roles for MLH Subunits in DNA Repair.Cells. 2021 Apr 20;10(4):948. doi: 10.3390/cells10040948. Cells. 2021. PMID: 33923939 Free PMC article. Review.
Cited by
-
Conditional knockdown of OsMLH1 to improve plant prime editing systems without disturbing fertility in rice.Genome Biol. 2024 May 21;25(1):131. doi: 10.1186/s13059-024-03282-y. Genome Biol. 2024. PMID: 38773623 Free PMC article.
-
Crossover interference mechanism: New lessons from plants.Front Cell Dev Biol. 2023 May 19;11:1156766. doi: 10.3389/fcell.2023.1156766. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37274744 Free PMC article. Review.
References
-
- Alou AH, Jean M, Domingue O, Belzile FJ (2004) Structure and expression of AtPMS1, the Arabidopsis ortholog of the yeast DNA repair gene PMS1. Plant Sci 167: 447–456
-
- Boddy MN, Gaillard PHL, McDonald WH, Shanahan P, Yates JR, Russell P (2001) Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548 - PubMed
-
- Cannavo E, Sanchez A, Anand R, Ranjha L, Hugener J, Adam C, Acharya A, Weyland N, Aran-Guiu X, Charbonnier JB, et al. (2020) Regulation of the MLH1-MLH3 endonuclease in meiosis. Nature 586: 618–622 - PubMed
-
- Cheng ZK (2013) Analyzing meiotic chromosomes in rice. Methods Mol Biol 990: 125–134 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

