Patterns of pan-genome occupancy and gene coexpression under water-deficit in Brachypodium distachyon
- PMID: 35976181
- PMCID: PMC9804473
- DOI: 10.1111/mec.16661
Patterns of pan-genome occupancy and gene coexpression under water-deficit in Brachypodium distachyon
Abstract
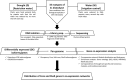
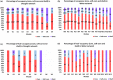
Natural populations are characterized by abundant genetic diversity driven by a range of different types of mutation. The tractability of sequencing complete genomes has allowed new insights into the variable composition of genomes, summarized as a species pan-genome. These analyses demonstrate that many genes are absent from the first reference genomes, whose analysis dominated the initial years of the genomic era. Our field now turns towards understanding the functional consequence of these highly variable genomes. Here, we analysed weighted gene coexpression networks from leaf transcriptome data for drought response in the purple false brome Brachypodium distachyon and the differential expression of genes putatively involved in adaptation to this stressor. We specifically asked whether genes with variable "occupancy" in the pan-genome - genes which are either present in all studied genotypes or missing in some genotypes - show different distributions among coexpression modules. Coexpression analysis united genes expressed in drought-stressed plants into nine modules covering 72 hub genes (87 hub isoforms), and genes expressed under controlled water conditions into 13 modules, covering 190 hub genes (251 hub isoforms). We find that low occupancy pan-genes are under-represented among several modules, while other modules are over-enriched for low-occupancy pan-genes. We also provide new insight into the regulation of drought response in B. distachyon, specifically identifying one module with an apparent role in primary metabolism that is strongly responsive to drought. Our work shows the power of integrating pan-genomic analysis with transcriptomic data using factorial experiments to understand the functional genomics of environmental response.
Keywords: genomics/proteomics; molecular evolution; phenotypic plasticity; transcriptomics.
© 2022 The Authors. Molecular Ecology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors affirm that they have no conflict of interest arising from this research or the publication of this manuscript.
Figures

Similar articles
-
Protocol for Coexpression Network Construction and Stress-Responsive Expression Analysis in Brachypodium.Methods Mol Biol. 2018;1667:203-221. doi: 10.1007/978-1-4939-7278-4_16. Methods Mol Biol. 2018. PMID: 29039014
-
Drought-inducible changes in the histone modification H3K9ac are associated with drought-responsive gene expression in Brachypodium distachyon.Plant Biol (Stuttg). 2020 May;22(3):433-440. doi: 10.1111/plb.13057. Epub 2020 Mar 11. Plant Biol (Stuttg). 2020. PMID: 31628708
-
Expression atlas and comparative coexpression network analyses reveal important genes involved in the formation of lignified cell wall in Brachypodium distachyon.New Phytol. 2017 Aug;215(3):1009-1025. doi: 10.1111/nph.14635. Epub 2017 Jun 15. New Phytol. 2017. PMID: 28617955
-
Prospects of pan-genomics in barley.Theor Appl Genet. 2019 Mar;132(3):785-796. doi: 10.1007/s00122-018-3234-z. Epub 2018 Nov 16. Theor Appl Genet. 2019. PMID: 30446793 Review.
-
Pan-genomics: Insight into the Functional Genome, Applications, Advancements, and Challenges.Curr Genomics. 2025;26(1):2-14. doi: 10.2174/0113892029311541240627111506. Epub 2024 Jul 3. Curr Genomics. 2025. PMID: 39911277 Free PMC article. Review.
Cited by
-
Comparative Genomics, Evolution, and Drought-Induced Expression of Dehydrin Genes in Model Brachypodium Grasses.Plants (Basel). 2021 Dec 3;10(12):2664. doi: 10.3390/plants10122664. Plants (Basel). 2021. PMID: 34961135 Free PMC article.
-
Transcriptomic and co-expression network analyses on diverse wheat landraces identifies candidate master regulators of the response to early drought.Front Plant Sci. 2023 Jun 23;14:1212559. doi: 10.3389/fpls.2023.1212559. eCollection 2023. Front Plant Sci. 2023. PMID: 37426985 Free PMC article.
References
-
- Alonge, M. , Wang, X. , Benoit, M. , Soyk, S. , Pereira, L. , Zhang, L. , Suresh, H. , Ramakrishnan, S. , Maumus, F. , Ciren, D. , Levy, Y. , Harel, T. H. , Shalev‐Schlosser, G. , Amsellem, Z. , Razifard, H. , Caicedo, A. L. , Tieman, D. M. , Klee, H. , Kirsche, M. , … Lippman, Z. B. (2020). Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell, 182(1), 145–161.e23. 10.1016/j.cell.2020.05.021 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

