Shell microelectrode arrays (MEAs) for brain organoids
- PMID: 35977026
- PMCID: PMC9385157
- DOI: 10.1126/sciadv.abq5031
Shell microelectrode arrays (MEAs) for brain organoids
Abstract
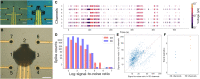
Brain organoids are important models for mimicking some three-dimensional (3D) cytoarchitectural and functional aspects of the brain. Multielectrode arrays (MEAs) that enable recording and stimulation of activity from electrogenic cells offer notable potential for interrogating brain organoids. However, conventional MEAs, initially designed for monolayer cultures, offer limited recording contact area restricted to the bottom of the 3D organoids. Inspired by the shape of electroencephalography caps, we developed miniaturized wafer-integrated MEA caps for organoids. The optically transparent shells are composed of self-folding polymer leaflets with conductive polymer-coated metal electrodes. Tunable folding of the minicaps' polymer leaflets guided by mechanics simulations enables versatile recording from organoids of different sizes, and we validate the feasibility of electrophysiology recording from 400- to 600-μm-sized organoids for up to 4 weeks and in response to glutamate stimulation. Our studies suggest that 3D shell MEAs offer great potential for high signal-to-noise ratio and 3D spatiotemporal brain organoid recording.
Figures




References
-
- Hartung T., Thoughts on limitations of animal models. Parkinsonism Relat. Disord. 14, S81–S83 (2008). - PubMed
-
- Trujillo C. A., Gao R., Negraes P. D., Gu J., Buchanan J., Preissl S., Wang A., Wu W., Haddad G. G., Chaim I. A., Domissy A., Vandenberghe M., Devor A., Yeo G. W., Voytek B., Muotri A. R., Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell 25, 558–569.e7 (2019). - PMC - PubMed
-
- Marx U., Akabane T., Andersson T. B., Baker E., Beilmann M., Beken S., Brendler-Schwaab S., Cirit M., David R., Dehne E. M., Durieux I., Ewart L., Fitzpatrick S. C., Frey O., Fuchs F., Griffith L. G., Hamilton G. A., Hartung T., Hoeng J., Hogberg H., Hughes D. J., Ingber D. E., Iskandar A., Kanamori T., Kojima H., Kuehnl J., Leist M., Li B., Loskill P., Mendrick D. L., Neumann T., Pallocca G., Rusyn I., Smirnova L., Steger-Hartmann T., Tagle D. A., Tonevitsky A., Tsyb S., Trapecar M., van de Water B., van den Eijnden-van Raaij J., Vulto P., Watanabe K., Wolf A., Zhou X., Roth A., Biology-inspired microphysiological systems to advance patient benefit and animal welfare in drug development. ALTEX 37, 365–394 (2020). - PMC - PubMed
-
- Roth A., Human microphysiological systems for drug development. Science 373, 1304–1306 (2021). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

