Discovery of novel predisposing coding and noncoding variants in familial Hodgkin lymphoma
- PMID: 35977101
- PMCID: PMC10082357
- DOI: 10.1182/blood.2022016056
Discovery of novel predisposing coding and noncoding variants in familial Hodgkin lymphoma
Abstract
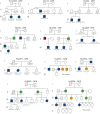
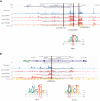
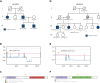
Familial aggregation of Hodgkin lymphoma (HL) has been demonstrated in large population studies, pointing to genetic predisposition to this hematological malignancy. To understand the genetic variants associated with the development of HL, we performed whole genome sequencing on 234 individuals with and without HL from 36 pedigrees that had 2 or more first-degree relatives with HL. Our pedigree selection criteria also required at least 1 affected individual aged <21 years, with the median age at diagnosis of 21.98 years (3-55 years). Family-based segregation analysis was performed for the identification of coding and noncoding variants using linkage and filtering approaches. Using our tiered variant prioritization algorithm, we identified 44 HL-risk variants in 28 pedigrees, of which 33 are coding and 11 are noncoding. The top 4 recurrent risk variants are a coding variant in KDR (rs56302315), a 5' untranslated region variant in KLHDC8B (rs387906223), a noncoding variant in an intron of PAX5 (rs147081110), and another noncoding variant in an intron of GATA3 (rs3824666). A newly identified splice variant in KDR (c.3849-2A>C) was observed for 1 pedigree, and high-confidence stop-gain variants affecting IRF7 (p.W238∗) and EEF2KMT (p.K116∗) were also observed. Multiple truncating variants in POLR1E were found in 3 independent pedigrees as well. Whereas KDR and KLHDC8B have previously been reported, PAX5, GATA3, IRF7, EEF2KMT, and POLR1E represent novel observations. Although there may be environmental factors influencing lymphomagenesis, we observed segregation of candidate germline variants likely to predispose HL in most of the pedigrees studied.
© 2023 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




Comment in
-
Unraveling family ties in Hodgkin lymphoma.Blood. 2023 Mar 16;141(11):1240-1241. doi: 10.1182/blood.2022018076. Blood. 2023. PMID: 36929435 No abstract available.
Similar articles
-
Identification of Familial Hodgkin Lymphoma Predisposing Genes Using Whole Genome Sequencing.Front Bioeng Biotechnol. 2020 Mar 6;8:179. doi: 10.3389/fbioe.2020.00179. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32211398 Free PMC article.
-
Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene.Haematologica. 2016 Jul;101(7):853-60. doi: 10.3324/haematol.2015.135475. Epub 2016 Jun 13. Haematologica. 2016. PMID: 27365461 Free PMC article.
-
Identification of Rare Variants Predisposing to Thyroid Cancer.Thyroid. 2019 Jul;29(7):946-955. doi: 10.1089/thy.2018.0736. Epub 2019 May 13. Thyroid. 2019. PMID: 30957677 Free PMC article.
-
Cancer predisposition and germline CTNNA1 variants.Eur J Med Genet. 2021 Oct;64(10):104316. doi: 10.1016/j.ejmg.2021.104316. Epub 2021 Aug 21. Eur J Med Genet. 2021. PMID: 34425242
-
Functional damaging germline variants in ETV6, IKZF1, PAX5 and RUNX1 predisposing to B-cell precursor acute lymphoblastic leukemia.Eur J Med Genet. 2023 Apr;66(4):104725. doi: 10.1016/j.ejmg.2023.104725. Epub 2023 Feb 9. Eur J Med Genet. 2023. PMID: 36764385 Review.
Cited by
-
Re-envisioning genetic predisposition to childhood and adolescent cancers.Nat Rev Cancer. 2025 Feb;25(2):109-128. doi: 10.1038/s41568-024-00775-7. Epub 2024 Dec 3. Nat Rev Cancer. 2025. PMID: 39627375 Review.
-
Children's Oncology Group's 2023 blueprint for research: Hodgkin lymphoma.Pediatr Blood Cancer. 2023 Sep;70 Suppl 6(Suppl 6):e30580. doi: 10.1002/pbc.30580. Epub 2023 Jul 28. Pediatr Blood Cancer. 2023. PMID: 37505794 Free PMC article.
-
Deep Learning in Hematology: From Molecules to Patients.Clin Hematol Int. 2024 Oct 8;6(4):19-42. doi: 10.46989/001c.124131. eCollection 2024. Clin Hematol Int. 2024. PMID: 39417017 Free PMC article. Review.
-
A Comprehensive Two-Decade Analysis of Lymphoma Incidence Patterns in Saudi Arabia.J Clin Med. 2024 Mar 13;13(6):1652. doi: 10.3390/jcm13061652. J Clin Med. 2024. PMID: 38541877 Free PMC article.
-
Investigating the tissue specificity and prognostic impact of cis-regulatory cancer risk variants.Hum Genet. 2023 Sep;142(9):1395-1405. doi: 10.1007/s00439-023-02586-6. Epub 2023 Jul 20. Hum Genet. 2023. PMID: 37474751
References
-
- Caporaso NE, Goldin LR, Anderson WF, Landgren O. Current insight on trends, causes, and mechanisms of Hodgkin's lymphoma. Cancer J. 2009;15(2):117–123. - PubMed
-
- Hidalgo J, Gasull T, Giralt M, Armario A. Brain metallothionein in stress. Biol Signals. 1994;3(4):198–210. - PubMed
-
- Mack TM, Cozen W, Shibata DK, et al. Concordance for Hodgkin's disease in identical twins suggesting genetic susceptibility to the young-adult form of the disease. N Engl J Med. 1995;332(7):413–418. - PubMed
-
- Kharazmi E, Fallah M, Pukkala E, et al. Risk of familial classical Hodgkin lymphoma by relationship, histology, age, and sex: a joint study from five Nordic countries. Blood. 2015;126(17):1990–1995. - PubMed
-
- Kuppers R. The biology of Hodgkin's lymphoma. Nat Rev Cancer. 2009;9(1):15–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

