Bergerac strains of Caenorhabditis elegans revisited: expansion of Tc1 elements imposes a significant genomic and fitness cost
- PMID: 35977391
- PMCID: PMC9635669
- DOI: 10.1093/g3journal/jkac214
Bergerac strains of Caenorhabditis elegans revisited: expansion of Tc1 elements imposes a significant genomic and fitness cost
Abstract
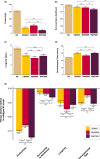
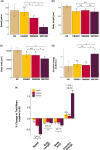
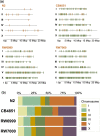
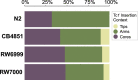
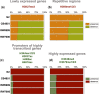
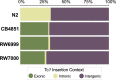
The DNA transposon Tc1 was the first transposable element to be characterized in Caenorhabditis elegans and to date, remains the best-studied transposable element in Caenorhabditis worms. While Tc1 copy-number is regulated at approximately 30 copies in the laboratory Bristol N2 and the vast majority of C. elegans strains, the Bergerac strain and its derivatives have experienced a marked Tc1 proliferation. Given the historical importance of the Bergerac strain in the development of the C. elegans model, we implemented a modern genomic analysis of three Bergerac strains (CB4851, RW6999, and RW7000) in conjunction with multiple phenotypic assays to better elucidate the (1) genomic distribution of Tc1 and (2) phenotypic consequences of transposable element deregulation for the host organism. The median estimates of Tc1 copy-number in the Bergerac strains ranged from 451 to 748, which is both (1) greater than previously estimated and (2) likely to be an underestimate of the actual copy-numbers since coverage-based estimates and digital droplet polymerase chain reaction results both suggest higher Tc1 numbers. All three Bergerac strains had significantly reduced trait means compared with the N2 control for each of four fitness-related traits, with specific traits displaying significant differences between Bergerac strains. Tc1 proliferation was genome-wide, specific to Tc1, and particularly high on chromosomes V and X. There were fewer Tc1 insertions in highly expressed chromatin environments than expected by chance. Furthermore, Tc1 integration motifs were also less frequent in exon than noncoding sequences. The source of the proliferation of Tc1 in the Bergerac strains is specific to Tc1 and independent of other transposable elements. The Bergerac strains contain none of the alleles that have previously been found to derepress transposable element activity in C. elegans. However, the Bergerac strains had several Tc1 insertions near or within highly germline-transcribed genes which could account for the recent germline proliferation.
Keywords: Caenorhabditis elegans; Bergerac; chromatin; copy-number variation; fitness; recombination; transposable element; transposon; whole-genome sequencing.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures
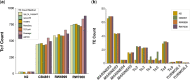

Similar articles
-
Tc1(Hin): a form of the transposable element Tc1 in Caenorhabditis elegans.Can J Biochem Cell Biol. 1985 Jul;63(7):752-6. doi: 10.1139/o85-094. Can J Biochem Cell Biol. 1985. PMID: 2994862
-
Evidence for a transposon in Caenorhabditis elegans.Cell. 1983 Jan;32(1):55-65. doi: 10.1016/0092-8674(83)90496-8. Cell. 1983. PMID: 6297788
-
Isolation of the closed circular form of the transposable element Tc1 in Caenorhabditis elegans.Nature. 1984 Oct 4-10;311(5985):485-6. doi: 10.1038/311485a0. Nature. 1984. PMID: 6090946
-
Towards a mutation in every gene in Caenorhabditis elegans.Brief Funct Genomic Proteomic. 2008 May;7(3):195-204. doi: 10.1093/bfgp/eln016. Epub 2008 Apr 16. Brief Funct Genomic Proteomic. 2008. PMID: 18417533 Review.
-
Transposons in C. elegans.WormBook. 2006 Jan 18:1-13. doi: 10.1895/wormbook.1.70.1. WormBook. 2006. PMID: 18023126 Free PMC article. Review.
Cited by
-
Life history in Caenorhabditis elegans: from molecular genetics to evolutionary ecology.Genetics. 2024 Nov 6;228(3):iyae151. doi: 10.1093/genetics/iyae151. Genetics. 2024. PMID: 39422376 Free PMC article. Review.
-
Caenorhabditis elegans foraging patterns follow a simple rule of thumb.Commun Biol. 2023 Aug 14;6(1):841. doi: 10.1038/s42003-023-05220-3. Commun Biol. 2023. PMID: 37580527 Free PMC article.
-
Reproducible evaluation of transposable element detectors with McClintock 2 guides accurate inference of Ty insertion patterns in yeast.bioRxiv [Preprint]. 2023 Mar 21:2023.02.13.528343. doi: 10.1101/2023.02.13.528343. bioRxiv. 2023. Update in: Mob DNA. 2023 Jul 14;14(1):8. doi: 10.1186/s13100-023-00296-4. PMID: 36824955 Free PMC article. Updated. Preprint.
-
Reproducible evaluation of transposable element detectors with McClintock 2 guides accurate inference of Ty insertion patterns in yeast.Mob DNA. 2023 Jul 14;14(1):8. doi: 10.1186/s13100-023-00296-4. Mob DNA. 2023. PMID: 37452430 Free PMC article.
References
-
- Abdulkader N, Brun J.. A temperature-sensitive mutant of Caenorhabditis elegans var. Bergerac affecting morphological and embryonic development. Genetics. 1980;51(2):81–92. doi:10.1007/bf00133506. - DOI
-
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D.. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13(10):807–818. doi:10.1016/s0960- 9822(03)00287-2. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
