A single-cell transcriptional gradient in human cutaneous memory T cells restricts Th17/Tc17 identity
- PMID: 35977472
- PMCID: PMC9418858
- DOI: 10.1016/j.xcrm.2022.100715
A single-cell transcriptional gradient in human cutaneous memory T cells restricts Th17/Tc17 identity
Abstract
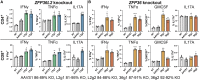
The homeostatic mechanisms that fail to restrain chronic tissue inflammation in diseases, such as psoriasis vulgaris, remain incompletely understood. We profiled transcriptomes and epitopes of single psoriatic and normal skin-resident T cells, revealing a gradated transcriptional program of coordinately regulated inflammation-suppressive genes. This program, which is sharply suppressed in lesional skin, strikingly restricts Th17/Tc17 cytokine and other inflammatory mediators on the single-cell level. CRISPR-based deactivation of two core components of this inflammation-suppressive program, ZFP36L2 and ZFP36, replicates the interleukin-17A (IL-17A), granulocyte macrophage-colony-stimulating factor (GM-CSF), and interferon gamma (IFNγ) elevation in psoriatic memory T cells deficient in these transcripts, functionally validating their influence. Combinatoric expression analysis indicates the suppression of specific inflammatory mediators by individual program members. Finally, we find that therapeutic IL-23 blockade reduces Th17/Tc17 cell frequency in lesional skin but fails to normalize this inflammatory-suppressive program, suggesting how treated lesions may be primed for recurrence after withdrawal of treatment.
Keywords: ZFP36; ZFP36L2; cytokine; inflammation; psoriasis; resident-memory T cell; tristetraprolin scRNA-seq.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.M. is a cofounder of Arsenal Biosciences, Spotlight Therapeutics, and Survey Genomics. A.M. serves on the boards of directors at Spotlight Therapeutics and Survey Genomics and is board observer (and former member of the board of directors) at Arsenal Biosciences. A.M. is a member of the scientific advisory boards of Arsenal Biosciences, Spotlight Therapeutics, Survey Genomics, and NewLimit. A.M. owns stock in Arsenal Biosciences, Spotlight Therapeutics, NewLimit, Survey Genomics, PACT Pharma, and Merck. A.M. has received fees from Arsenal Biosciences, Spotlight Therapeutics, NewLimit, 23andMe, PACT Pharma, Juno Therapeutics, Trizell, Vertex, Merck, Amgen, Genentech, AlphaSights, Rupert Case Management, Bernstein, and ALDA. A.M. is an investor in and informal advisor to Offline Ventures and a client of EPIQ.
Figures





References
-
- Lowes M.A., Kikuchi T., Fuentes-Duculan J., Cardinale I., Zaba L.C., Haider A.S., Bowman E.P., Krueger J.G. Psoriasis vulgaris lesions contain discrete populations of Th1 and Th17 T cells. J. Invest. Dermatol. 2008;128:1207–1211. - PubMed
-
- Hughes T.K., Wadsworth M.H., Gierahn T.M., Do T., Weiss D., Andrade P.R., Ma F., de Andrade Silva B.J., Shao S., Tsoi L.C., et al. Second-strand synthesis-based Massively Parallel scRNA-seq reveals cellular states and molecular features of human inflammatory skin Pathologies. Immunity. 2020;53:878–894.e7. - PMC - PubMed

