NF-κB memory coordinates transcriptional responses to dynamic inflammatory stimuli
- PMID: 35977475
- PMCID: PMC10794069
- DOI: 10.1016/j.celrep.2022.111159
NF-κB memory coordinates transcriptional responses to dynamic inflammatory stimuli
Abstract
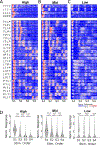
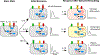
Many scenarios in cellular communication require cells to interpret multiple dynamic signals. It is unclear how exposure to inflammatory stimuli alters transcriptional responses to subsequent stimulus. Using high-throughput microfluidic live-cell analysis, we systematically profile the NF-κB response to different signal sequences in single cells. We find that NF-κB dynamics store the short-term history of received signals: depending on the prior pathogenic or cytokine signal, the NF-κB response to subsequent stimuli varies from no response to full activation. Using information theory, we reveal that these stimulus-dependent changes in the NF-κB response encode and reflect information about the identity and dose of the prior stimulus. Small-molecule inhibition, computational modeling, and gene expression profiling show that this encoding is driven by stimulus-dependent engagement of negative feedback modules. These results provide a model for how signal transduction networks process sequences of inflammatory stimuli to coordinate cellular responses in complex dynamic environments.
Keywords: CP: Immunology; NF-κB; cellular memory; inflammation; information theory; innate immune signaling; live-cell imaging; mathematical modeling; microfluidics; pathogen-associated molecular patterns; signaling dynamics.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Adamson A, Boddington C, Downton P, Rowe W, Bagnall J, Lam C, Maya-Mendoza A, Schmidt L, Harper CV, Spiller DG, et al. (2016). Signal transduction controls heterogeneous NF-κB dynamics and target gene expression through cytokine-specific refractory states. Nat Commun 7, 12057. 10.1038/ncomms12057. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

