341 Repeats Is Not Enough for Methylation in a New Fragile X Mouse Model
- PMID: 35977823
- PMCID: PMC9469916
- DOI: 10.1523/ENEURO.0142-22.2022
341 Repeats Is Not Enough for Methylation in a New Fragile X Mouse Model
Abstract
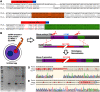
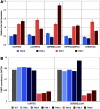
Fragile X syndrome (FXS) is a leading monogenic cause of intellectual disability and autism spectrum disorders, spurring decades of intense research and a multitude of mouse models. So far, these models do not recapitulate the genetic underpinning of classical FXS-CGG repeat-induced methylation of the Fmr1 locus-and their findings have failed to translate into the clinic. We sought to answer whether this disparity was because of low repeat length and generated a novel mouse line with 341 repeats, Fmr1hs341 , which is the largest allele in mice reported to date. This repeat length is significantly longer than the 200 repeats generally required for methylation of the repeat tract and promoter region in FXS patients, which leads to silencing of the FMR1 gene. Bisulfite sequencing fails to detect the robust methylation expected of FXS in Fmr1hs341 mice. Quantitative real-time PCR and Western blotting results also do not resemble FXS and instead produce a biochemical profile consistent with the fragile X-associated premutation disorders. These findings suggest that repeat length is unlikely to be the core determinant preventing methylation in mice, and other organisms phylogenetically closer to humans may be required to effectively model FXS.
Copyright © 2022 Colvin et al.
Figures

Similar articles
-
Transcriptomic profiling of unmethylated full mutation carriers implicates TET3 in FMR1 CGG repeat expansion methylation dynamics in fragile X syndrome.J Neurodev Disord. 2025 Apr 26;17(1):22. doi: 10.1186/s11689-025-09609-5. J Neurodev Disord. 2025. PMID: 40287634 Free PMC article.
-
CGG-repeat dynamics and FMR1 gene silencing in fragile X syndrome stem cells and stem cell-derived neurons.Mol Autism. 2016 Oct 6;7:42. doi: 10.1186/s13229-016-0105-9. eCollection 2016. Mol Autism. 2016. PMID: 27713816 Free PMC article.
-
Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome.PLoS One. 2011;6(10):e26203. doi: 10.1371/journal.pone.0026203. Epub 2011 Oct 12. PLoS One. 2011. PMID: 22022567 Free PMC article.
-
Phenotypic variability to medication management: an update on fragile X syndrome.Hum Genomics. 2023 Jul 7;17(1):60. doi: 10.1186/s40246-023-00507-2. Hum Genomics. 2023. PMID: 37420260 Free PMC article. Review.
-
Unstable mutations in the FMR1 gene and the phenotypes.Adv Exp Med Biol. 2012;769:78-114. doi: 10.1007/978-1-4614-5434-2_6. Adv Exp Med Biol. 2012. PMID: 23560306 Free PMC article. Review.
Cited by
-
Mouse models of fragile X-related disorders.Dis Model Mech. 2023 Feb 1;16(2):dmm049485. doi: 10.1242/dmm.049485. Epub 2023 Jan 24. Dis Model Mech. 2023. PMID: 36692473 Free PMC article. Review.
-
From wings to whiskers to stem cells: why every model matters in fragile X syndrome research.J Neurodev Disord. 2024 Jun 13;16(1):30. doi: 10.1186/s11689-024-09545-w. J Neurodev Disord. 2024. PMID: 38872088 Free PMC article. Review.
-
Transcriptomic profiling of unmethylated full mutation carriers implicates TET3 in FMR1 CGG repeat expansion methylation dynamics in fragile X syndrome.J Neurodev Disord. 2025 Apr 26;17(1):22. doi: 10.1186/s11689-025-09609-5. J Neurodev Disord. 2025. PMID: 40287634 Free PMC article.
-
Ascorbic Acid Ameliorates Molecular and Developmental Defects in Human-Induced Pluripotent Stem Cell and Cerebral Organoid Models of Fragile X Syndrome.Int J Mol Sci. 2024 Nov 26;25(23):12718. doi: 10.3390/ijms252312718. Int J Mol Sci. 2024. PMID: 39684429 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
