Promotion of neutralizing antibody-independent immunity to wild-type and SARS-CoV-2 variants of concern using an RBD-Nucleocapsid fusion protein
- PMID: 35977933
- PMCID: PMC9382605
- DOI: 10.1038/s41467-022-32547-y
Promotion of neutralizing antibody-independent immunity to wild-type and SARS-CoV-2 variants of concern using an RBD-Nucleocapsid fusion protein
Abstract
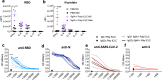
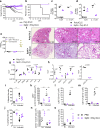
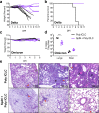
Both T cells and B cells have been shown to be generated after infection with SARS-CoV-2 yet protocols or experimental models to study one or the other are less common. Here, we generate a chimeric protein (SpiN) that comprises the receptor binding domain (RBD) from Spike (S) and the nucleocapsid (N) antigens from SARS-CoV-2. Memory CD4+ and CD8+ T cells specific for SpiN could be detected in the blood of both individuals vaccinated with Coronavac SARS-CoV-2 vaccine and COVID-19 convalescent donors. In mice, SpiN elicited a strong IFN-γ response by T cells and high levels of antibodies to the inactivated virus, but not detectable neutralizing antibodies (nAbs). Importantly, immunization of Syrian hamsters and the human Angiotensin Convertase Enzyme-2-transgenic (K18-ACE-2) mice with Poly ICLC-adjuvanted SpiN promotes robust resistance to the wild type SARS-CoV-2, as indicated by viral load, lung inflammation, clinical outcome and reduction of lethality. The protection induced by SpiN was ablated by depletion of CD4+ and CD8+ T cells and not transferred by antibodies from vaccinated mice. Finally, vaccination with SpiN also protects the K18-ACE-2 mice against infection with Delta and Omicron SARS-CoV-2 isolates. Hence, vaccine formulations that elicit effector T cells specific for the N and RBD proteins may be used to improve COVID-19 vaccines and potentially circumvent the immune escape by variants of concern.
© 2022. The Author(s).
Conflict of interest statement
R.T.G., S.R.T., A.P.F., F.F., N.S., J.C., N.S.H-S., and P.A. are co-inventors of the potential COVID-19 vaccine evaluated in this study. The patent is under evaluation process, application number BR1020210095733. The other authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

