LINE-1 and Alu methylation signatures in autism spectrum disorder and their associations with the expression of autism-related genes
- PMID: 35978033
- PMCID: PMC9385849
- DOI: 10.1038/s41598-022-18232-6
LINE-1 and Alu methylation signatures in autism spectrum disorder and their associations with the expression of autism-related genes
Abstract
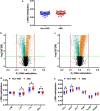
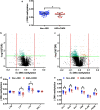
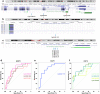
Long interspersed nucleotide element-1 (LINE-1) and Alu elements are retrotransposons whose abilities cause abnormal gene expression and genomic instability. Several studies have focused on DNA methylation profiling of gene regions, but the locus-specific methylation of LINE-1 and Alu elements has not been identified in autism spectrum disorder (ASD). Here we interrogated locus- and family-specific methylation profiles of LINE-1 and Alu elements in ASD whole blood using publicly-available Illumina Infinium 450 K methylation datasets from heterogeneous ASD and ASD variants (Chromodomain Helicase DNA-binding 8 (CHD8) and 16p11.2del). Total DNA methylation of repetitive elements were notably hypomethylated exclusively in ASD with CHD8 variants. Methylation alteration in a family-specific manner including L1P, L1H, HAL, AluJ, and AluS families were observed in the heterogeneous ASD and ASD with CHD8 variants. Moreover, LINE-1 and Alu methylation within target genes is inversely related to the expression level in each ASD variant. The DNA methylation signatures of the LINE-1 and Alu elements in ASD whole blood, as well as their associations with the expression of ASD-related genes, have been identified. If confirmed in future larger studies, these findings may contribute to the identification of epigenomic biomarkers of ASD.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Association AP. Diagnostic and Statistical Manual of Mental Disorders 5th edn (2013).
-
- Maenner MJ, Shaw KA, Bakian AV, Bilder DA, Durkin MS, Esler A, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and developmental disabilities monitoring network, 11 sites, United States, 2018. MMWR Surveill. Summ. 2021;70(11):1–16. doi: 10.15585/mmwr.ss7011a1. - DOI - PMC - PubMed

