Genome characterization and CRISPR-Cas9 editing of a human neocentromere
- PMID: 35978051
- PMCID: PMC9674717
- DOI: 10.1007/s00412-022-00779-y
Genome characterization and CRISPR-Cas9 editing of a human neocentromere
Abstract
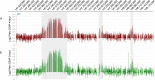
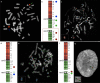
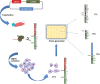
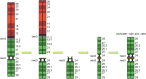
The maintenance of genome integrity is ensured by proper chromosome inheritance during mitotic and meiotic cell divisions. The chromosomal counterpart responsible for chromosome segregation to daughter cells is the centromere, at which the spindle apparatus attaches through the kinetochore. Although all mammalian centromeres are primarily composed of megabase-long repetitive sequences, satellite-free human neocentromeres have been described. Neocentromeres and evolutionary new centromeres have revolutionized traditional knowledge about centromeres. Over the past 20 years, insights have been gained into their organization, but in spite of these advancements, the mechanisms underlying their formation and evolution are still unclear. Today, through modern and increasingly accessible genome editing and long-read sequencing techniques, research in this area is undergoing a sudden acceleration. In this article, we describe the primary sequence of a previously described human chromosome 3 neocentromere and observe its possible evolution and repair results after a chromosome breakage induced through CRISPR-Cas9 technologies. Our data represent an exciting advancement in the field of centromere/neocentromere evolution and chromosome stability.
Keywords: CRISPR-Cas9; Isochromosome; Long-read sequencing; Neocentromere.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Neocentromeres Provide Chromosome Segregation Accuracy and Centromere Clustering to Multiple Loci along a Candida albicans Chromosome.PLoS Genet. 2016 Sep 23;12(9):e1006317. doi: 10.1371/journal.pgen.1006317. eCollection 2016 Sep. PLoS Genet. 2016. PMID: 27662467 Free PMC article.
-
Neocentromeres and epigenetically inherited features of centromeres.Chromosome Res. 2012 Jul;20(5):607-19. doi: 10.1007/s10577-012-9296-x. Chromosome Res. 2012. PMID: 22723125 Free PMC article. Review.
-
Formation of novel CENP-A domains on tandem repetitive DNA and across chromosome breakpoints on human chromosome 8q21 neocentromeres.Chromosoma. 2011 Dec;120(6):621-32. doi: 10.1007/s00412-011-0337-6. Epub 2011 Aug 9. Chromosoma. 2011. PMID: 21826412
-
Artificial generation of centromeres and kinetochores to understand their structure and function.Exp Cell Res. 2020 Apr 15;389(2):111898. doi: 10.1016/j.yexcr.2020.111898. Epub 2020 Feb 6. Exp Cell Res. 2020. PMID: 32035949 Review.
-
Epigenetic control of centromere: what can we learn from neocentromere?Genes Genomics. 2022 Mar;44(3):317-325. doi: 10.1007/s13258-021-01193-x. Epub 2021 Nov 29. Genes Genomics. 2022. PMID: 34843088 Review.
Cited by
-
Robertsonian Translocation between Human Chromosomes 21 and 22, Inherited across Three Generations, without Any Phenotypic Effect.Genes (Basel). 2024 Jun 1;15(6):722. doi: 10.3390/genes15060722. Genes (Basel). 2024. PMID: 38927657 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

