Computational studies indicated the effectiveness of human metabolites against SARS-Cov-2 main protease
- PMID: 35978064
- PMCID: PMC9385416
- DOI: 10.1007/s11030-022-10513-6
Computational studies indicated the effectiveness of human metabolites against SARS-Cov-2 main protease
Abstract
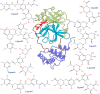
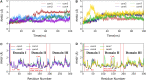
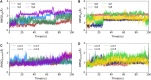
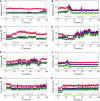
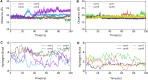
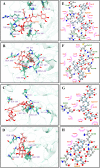
To fight against the devastating coronavirus disease 2019 (COVID-19), identifying robust anti-SARS-CoV-2 therapeutics from all possible directions is necessary. To contribute to this effort, we selected a human metabolites database containing waters and lipid-soluble metabolites to screen against the 3-chymotrypsin-like proteases (3CLpro) protein of SARS-CoV-2. The top 8 hits from virtual screening displayed a docking score varying between ~ - 11 and ~ - 14 kcal/mol. Molecular dynamics simulations complement the virtual screening study in conjunction with the molecular mechanics generalized Born surface area (MM/GBSA) scheme. Our analyses revealed that (HMDB0132640) has the best glide docking score, - 14.06 kcal/mol, and MM-GBSA binding free energy, - 18.08 kcal/mol. The other three lead molecules are also selected along with the top molecule through a critical inspection of their pharmacokinetic properties. HMDB0132640 displayed a better binding affinity than the other three compounds (HMDB0127868, HMDB0134119, and HMDB0125821) due to increased favorable contributions from the intermolecular electrostatic and van der Waals interactions. Further, we have investigated the ligand-induced structural dynamics of the main protease. Overall, we have identified new compounds that can serve as potential leads for developing novel antiviral drugs against SARS-CoV-2 and elucidated molecular mechanisms of their binding to the main protease. Identification of probable hits from human metabolites against SARS-CoV-2 using integrated computational approaches-Missed against MS.
Keywords: Free energy; Human metabolites; Main protease; Molecular dynamics; SARS-COV-2; Virtual screening.
© 2022. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

References
-
- Madboly WE, Shehata MG, Nashed MSM, Abu-Dief AM. Using safe calculated low power of electrons to cut, analyze and exterminate the outer and inner biological elements of SARS-CoV-2, MERS-CoV-2 and influenza viruses in vitro. J Sci Res Rep. 2022;28(1):1–15. doi: 10.9734/jsrr/2022/v28i130482. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

