Endocytosis in the axon initial segment maintains neuronal polarity
- PMID: 35978188
- PMCID: PMC9433327
- DOI: 10.1038/s41586-022-05074-5
Endocytosis in the axon initial segment maintains neuronal polarity
Erratum in
-
Author Correction: Endocytosis in the axon initial segment maintains neuronal polarity.Nature. 2025 Apr;640(8057):E2. doi: 10.1038/s41586-025-08837-y. Nature. 2025. PMID: 40102666 Free PMC article. No abstract available.
Abstract
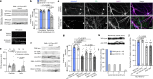
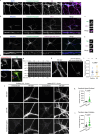
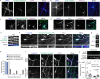
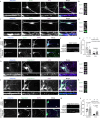
Neurons are highly polarized cells that face the fundamental challenge of compartmentalizing a vast and diverse repertoire of proteins in order to function properly1. The axon initial segment (AIS) is a specialized domain that separates a neuron's morphologically, biochemically and functionally distinct axon and dendrite compartments2,3. How the AIS maintains polarity between these compartments is not fully understood. Here we find that in Caenorhabditis elegans, mouse, rat and human neurons, dendritically and axonally polarized transmembrane proteins are recognized by endocytic machinery in the AIS, robustly endocytosed and targeted to late endosomes for degradation. Forcing receptor interaction with the AIS master organizer, ankyrinG, antagonizes receptor endocytosis in the AIS, causes receptor accumulation in the AIS, and leads to polarity deficits with subsequent morphological and behavioural defects. Therefore, endocytic removal of polarized receptors that diffuse into the AIS serves as a membrane-clearance mechanism that is likely to work in conjunction with the known AIS diffusion-barrier mechanism to maintain neuronal polarity on the plasma membrane. Our results reveal a conserved endocytic clearance mechanism in the AIS to maintain neuronal polarity by reinforcing axonal and dendritic compartment membrane boundaries.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- Bentley, M. & Banker, G. The cellular mechanisms that maintain neuronal polarity. Nat. Rev. Neurosci.17, 611–622 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

