Flexibility of flanking DNA is a key determinant of transcription factor affinity for the core motif
- PMID: 35978548
- PMCID: PMC9674967
- DOI: 10.1016/j.bpj.2022.08.015
Flexibility of flanking DNA is a key determinant of transcription factor affinity for the core motif
Abstract
Selective gene regulation is mediated by recognition of specific DNA sequences by transcription factors (TFs). The extremely challenging task of searching out specific cognate DNA binding sites among several million putative sites within the eukaryotic genome is achieved by complex molecular recognition mechanisms. Elements of this recognition code include the core binding sequence, the flanking sequence context, and the shape and conformational flexibility of the composite binding site. To unravel the extent to which DNA flexibility modulates TF binding, in this study, we employed experimentally guided molecular dynamics simulations of ternary complex of closely related Hox heterodimers Exd-Ubx and Exd-Scr with DNA. Results demonstrate that flexibility signatures embedded in the flanking sequences impact TF binding at the cognate binding site. A DNA sequence has intrinsic shape and flexibility features. While shape features are localized, our analyses reveal that flexibility features of the flanking sequences percolate several basepairs and allosterically modulate TF binding at the core. We also show that lack of flexibility in the motif context can render the cognate site resistant to protein-induced shape changes and subsequently lower TF binding affinity. Overall, this study suggests that flexibility-guided DNA shape, and not merely the static shape, is a key unexplored component of the complex DNA-TF recognition code.
Copyright © 2022 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
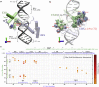


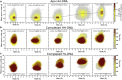

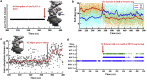

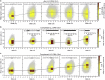
Comment in
-
It is in the flanks: Conformational flexibility of transcription factor binding sites.Biophys J. 2022 Oct 18;121(20):3765-3767. doi: 10.1016/j.bpj.2022.09.020. Epub 2022 Sep 21. Biophys J. 2022. PMID: 36182667 Free PMC article. No abstract available.
Similar articles
-
Probing the role of the protonation state of a minor groove-linker histidine in Exd-Hox-DNA binding.Biophys J. 2024 Jan 16;123(2):248-259. doi: 10.1016/j.bpj.2023.12.013. Epub 2023 Dec 21. Biophys J. 2024. PMID: 38130056 Free PMC article.
-
Flexibility and structure of flanking DNA impact transcription factor affinity for its core motif.Nucleic Acids Res. 2018 Dec 14;46(22):11883-11897. doi: 10.1093/nar/gky1057. Nucleic Acids Res. 2018. PMID: 30395339 Free PMC article.
-
Discerning the Role of DNA Sequence, Shape, and Flexibility in Recognition by Drosophila Transcription Factors.ACS Chem Biol. 2024 Jul 19;19(7):1533-1543. doi: 10.1021/acschembio.4c00202. Epub 2024 Jun 20. ACS Chem Biol. 2024. PMID: 38902964
-
How motif environment influences transcription factor search dynamics: Finding a needle in a haystack.Bioessays. 2016 Jul;38(7):605-12. doi: 10.1002/bies.201600005. Epub 2016 May 19. Bioessays. 2016. PMID: 27192961 Free PMC article. Review.
-
Towards a better understanding of TF-DNA binding prediction from genomic features.Comput Biol Med. 2022 Oct;149:105993. doi: 10.1016/j.compbiomed.2022.105993. Epub 2022 Aug 17. Comput Biol Med. 2022. PMID: 36057196 Review.
Cited by
-
Modeling and designing enhancers by introducing and harnessing transcription factor binding units.Nat Commun. 2025 Feb 8;16(1):1469. doi: 10.1038/s41467-025-56749-2. Nat Commun. 2025. PMID: 39922842 Free PMC article.
-
A high throughput single molecule platform to study DNA supercoiling effect on protein-DNA interactions.bioRxiv [Preprint]. 2024 Oct 25:2024.10.24.620099. doi: 10.1101/2024.10.24.620099. bioRxiv. 2024. Update in: Nucleic Acids Res. 2025 Jun 20;53(12):gkaf581. doi: 10.1093/nar/gkaf581. PMID: 39484392 Free PMC article. Updated. Preprint.
-
Probing the role of the protonation state of a minor groove-linker histidine in Exd-Hox-DNA binding.Biophys J. 2024 Jan 16;123(2):248-259. doi: 10.1016/j.bpj.2023.12.013. Epub 2023 Dec 21. Biophys J. 2024. PMID: 38130056 Free PMC article.
-
Variations in flanking or less conserved positions of Reb1 and Abf1 consensus binding sites lead to major changes in their ability to modulate nucleosome sliding activity.Biol Res. 2025 Jul 29;58(1):53. doi: 10.1186/s40659-025-00627-0. Biol Res. 2025. PMID: 40731031 Free PMC article.
-
Predictions of DNA mechanical properties at a genomic scale reveal potentially new functional roles of DNA flexibility.NAR Genom Bioinform. 2023 Nov 6;5(4):lqad097. doi: 10.1093/nargab/lqad097. eCollection 2023 Dec. NAR Genom Bioinform. 2023. PMID: 37954573 Free PMC article.
References
-
- Jones S., van Heyningen P., et al. Thornton J.M. Protein-DNA interactions: a structural analysis. J. Mol. Biol. 1999;287:877–896. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

