Fusarium and allied fusarioid taxa (FUSA). 1
- PMID: 35978986
- PMCID: PMC9355104
- DOI: 10.3114/fuse.2022.09.08
Fusarium and allied fusarioid taxa (FUSA). 1
Abstract
Seven Fusarium species complexes are treated, namely F. aywerte species complex (FASC) (two species), F. buharicum species complex (FBSC) (five species), F. burgessii species complex (FBURSC) (three species), F. camptoceras species complex (FCAMSC) (three species), F. chlamydosporum species complex (FCSC) (eight species), F. citricola species complex (FCCSC) (five species) and the F. concolor species complex (FCOSC) (four species). New species include Fusicolla elongata from soil (Zimbabwe), and Neocosmospora geoasparagicola from soil associated with Asparagus officinalis (Netherlands). New combinations include Neocosmospora akasia, N. awan, N. drepaniformis, N. duplosperma, N. geoasparagicola, N. mekan, N. papillata, N. variasi and N. warna. Newly validated taxa include Longinectria gen. nov., L. lagenoides, L. verticilliforme, Fusicolla gigas and Fusicolla guangxiensis. Furthermore, Fusarium rosicola is reduced to synonymy under N. brevis. Finally, the genome assemblies of Fusarium secorum (CBS 175.32), Microcera coccophila (CBS 310.34), Rectifusarium robinianum (CBS 430.91), Rugonectria rugulosa (CBS 126565), and Thelonectria blattea (CBS 952.68) are also announced here. Citation: Crous PW, Sandoval-Denis M, Costa MM, Groenewald JZ, van Iperen AL, Starink-Willemse M, Hernández-Restrepo M, Kandemir H, Ulaszewski B, de Boer W, Abdel-Azeem AM, Abdollahzadeh J, Akulov A, Bakhshi M, Bezerra JDP, Bhunjun CS, Câmara MPS, Chaverri P, Vieira WAS, Decock CA, Gaya E, Gené J, Guarro J, Gramaje D, Grube M, Gupta VK, Guarnaccia V, Hill R, Hirooka Y, Hyde KD, Jayawardena RS, Jeewon R, Jurjević Ž, Korsten L, Lamprecht SC, Lombard L, Maharachchikumbura SSN, Polizzi G, Rajeshkumar KC, Salgado-Salazar C, Shang Q-J, Shivas RG, Summerbell RC, Sun GY, Swart WJ, Tan YP, Vizzini A, Xia JW, Zare R, González CD, Iturriaga T, Savary O, Coton M, Coton E, Jany J-L, Liu C, Zeng Z-Q, Zhuang W-Y, Yu Z-H, Thines M (2022). Fusarium and allied fusarioid taxa (FUSA). 1. Fungal Systematics and Evolution 9: 161-200. doi: 10.3114/fuse.2022.09.08.
Keywords: Longinectria; Nectriaceae; Neocosmospora; multi-gene phylogeny; new taxa; systematics; typification.
© 2022 Westerdijk Fungal Biodiversity Institute.
Conflict of interest statement
Conflict of interest: The authors declare that there is no conflict of interest.
Figures
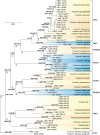














References
-
- Adamčík S, Cai L, Chakraborty D, et al. (2015). Fungal Biodiversity Profiles 1–10. Cryptogamie, Mycologie 36: 121–166.
-
- Ahmad A, Akram W, Shahzadi I, et al. (2020). First report of Fusarium nelsonii causing early-stage fruit blight of cucumber in Guangzhou, China. Plant Disease 104: 1542.
-
- Aoki T, Liyanage PNH, Konkol JL, et al. (2021). Three novel Ambrosia Fusarium Clade species producing multiseptate “dolphin-shaped” conidia, and an augmented description of Fusarium kuroshium. Mycologia 113: 1089–1109. - PubMed
-
- Aoki T, O’Donnell K, Geiser D. (2014). Systematics of key phytopathogenic Fusarium species: current status and future challenges. Journal of General Plant Pathology 80: 189–201.
-
- Balmas V, Migheli Q, Scherm B, et al. (2010). Multilocus phylogenetics show high levels of endemic fusaria inhabiting Sardinian soils (Tyrrhenian Islands). Mycologia 102: 803–812. - PubMed
LinkOut - more resources
Full Text Sources
