Novel multiplex stool-based assay for the detection of early-stage colon cancer in a Chinese population
- PMID: 35979157
- PMCID: PMC9260868
- DOI: 10.3748/wjg.v28.i24.2705
Novel multiplex stool-based assay for the detection of early-stage colon cancer in a Chinese population
Abstract
Background: Stool DNA (sDNA) methylation analysis is a promising, noninvasive approach for colorectal cancer screening; however, reliable biomarkers for detecting early-stage colon cancer (ECC) are lacking, particularly in the Chinese population.
Aim: To identify a novel stool-based assay that can improve the effectiveness of ECC screening.
Methods: A blinded case-control study was performed using archived stool samples from 125 ECC patients, and 125 control subjects with normal colonoscopy. The cohort was randomly divided into training and test sets at a 1.5:1 ratio. Targeted bisulfite sequencing (TBSeq) was conducted on five pairs of preoperative and postop-erative sDNA samples from ECC patients to identify DNA methylation biomarkers, which were validated using pyrosequencing. By logistic regression analysis, a multiplex stool-based assay was developed in the training set, and the detection performance was further assessed in the test set and combined set. The χ 2 test was used to investigate the association of detection sensitivity with clinico-pathological features.
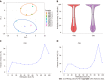
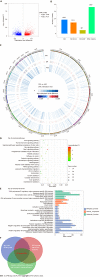
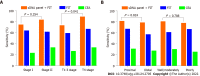
Results: Following TBSeq, three hypermethylated cytosine-guanine sites were selected as biomarkers, including paired box 8, Ras-association domain family 1 and secreted frizzled-related protein 2, which differed between the groups and were involved in important cancer pathways. An sDNA panel containing the three biomarkers was constructed with a logistic model. Receiver operating characteristic (ROC) analysis revealed that this panel was superior to the fecal immunochemical test (FIT) or serum carcinoembryonic antigen for the detection of ECC. We further found that the combination of the sDNA panel with FIT could improve the screening effectiveness. In the combined set, the sensitivity, specificity and area under the ROC curve for this multiplex assay were 80.0%, 93.6% and 0.918, respectively, and the performance remained excellent in the subgroup analysis by tumor stage. In addition, the detection sensitivity did not differ with tumor site, tumor stage, histological differentiation, age or sex, but was significantly higher in T4 than in T1-3 stage tumors (P = 0.041).
Conclusion: We identified a novel multiplex stool-based assay combining sDNA methylation biomarkers and FIT, which could detect ECC with high sensitivity and specificity throughout the colon, showing a promising application perspective.
Keywords: Colon cancer; DNA methylation; Early screening; Fecal immunochemical test; Stool biomarker.
©The Author(s) 2022. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors have no conflicts of interest to declare.
Figures


Similar articles
-
Robust performance of a novel stool DNA test of methylated SDC2 for colorectal cancer detection: a multicenter clinical study.Clin Epigenetics. 2020 Oct 30;12(1):162. doi: 10.1186/s13148-020-00954-x. Clin Epigenetics. 2020. PMID: 33126908 Free PMC article. Clinical Trial.
-
Multitarget Stool DNA Test Performance in an Average-Risk Colorectal Cancer Screening Population.Am J Gastroenterol. 2019 Dec;114(12):1909-1918. doi: 10.14309/ajg.0000000000000445. Am J Gastroenterol. 2019. PMID: 31764091 Free PMC article.
-
Analysis of the effectiveness of two noninvasive fecal tests used to screen for colorectal cancer in average-risk adults.Public Health. 2020 May;182:70-76. doi: 10.1016/j.puhe.2020.01.021. Epub 2020 Mar 13. Public Health. 2020. PMID: 32179290
-
Fecal DNA Testing for Colorectal Cancer Screening.Annu Rev Med. 2020 Jan 27;71:59-69. doi: 10.1146/annurev-med-103018-123125. Epub 2019 Aug 26. Annu Rev Med. 2020. PMID: 31451044 Review.
-
Noninvasive fecal testing for colorectal cancer.Clin Chim Acta. 2022 Jan 1;524:123-131. doi: 10.1016/j.cca.2021.10.030. Epub 2021 Oct 28. Clin Chim Acta. 2022. PMID: 34756863 Review.
Cited by
-
Identification of a PANoptosis-related long noncoding rna risk signature for prognosis and immunology in colon adenocarcinoma.BMC Cancer. 2025 Apr 10;25(1):662. doi: 10.1186/s12885-025-14021-2. BMC Cancer. 2025. PMID: 40211224 Free PMC article.
-
Early Detection, Precision Treatment, Recurrence Monitoring: Liquid Biopsy Transforms Colorectal Cancer Therapy.Curr Cancer Drug Targets. 2025;25(6):586-619. doi: 10.2174/0115680096295070240318075023. Curr Cancer Drug Targets. 2025. PMID: 38623975 Review.
-
Diagnostic value of genetic and epigenetic biomarker panels for colorectal cancer detection: a systematic review.Int J Colorectal Dis. 2025 May 22;40(1):125. doi: 10.1007/s00384-025-04904-y. Int J Colorectal Dis. 2025. PMID: 40402271 Free PMC article.
References
-
- Levin TR, Corley DA, Jensen CD, Schottinger JE, Quinn VP, Zauber AG, Lee JK, Zhao WK, Udaltsova N, Ghai NR, Lee AT, Quesenberry CP, Fireman BH, Doubeni CA. Effects of Organized Colorectal Cancer Screening on Cancer Incidence and Mortality in a Large Community-Based Population. Gastroenterology. 2018;155:1383–1391.e5. - PMC - PubMed
-
- Lou S, Shaukat A. Noninvasive strategies for colorectal cancer screening: opportunities and limitations. Curr Opin Gastroenterol. 2021;37:44–51. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

