Clonal serotype 1c multidrug-resistant Shigella flexneri detected in multiple institutions by sentinel-site sequencing
- PMID: 35979220
- PMCID: PMC9376355
- DOI: 10.3389/fmed.2022.964640
Clonal serotype 1c multidrug-resistant Shigella flexneri detected in multiple institutions by sentinel-site sequencing
Abstract
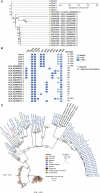
Shigella flexneri is a major diarrhoeal pathogen, and the emergence of multidrug-resistant S. flexneri is of public health concern. We report the detection of a clonal cluster of multidrug-resistant serotype 1c (7a) S. flexneri in Singapore in April 2022. Long-read whole-genome sequence analysis found five S. flexneri isolates to be clonal and harboring the extended-spectrum β-lactamases bla CTX-M-15 and bla TEM-1. The isolates were phenotypically resistant to ceftriaxone and had intermediate susceptibility to ciprofloxacin. The S. flexneri clonal cluster was first detected in a tertiary hospital diagnostic laboratory (sentinel-site), to which the S. flexneri isolates were sent from other hospitals for routine serogrouping. Long-read whole-genome sequence analysis was performed in the sentinel-site near real-time in view of the unusually high number of S. flexneri isolates received within a short time frame. This study demonstrates that near real-time sentinel-site sequence-based surveillance of convenience samples can detect possible clonal outbreak clusters and may provide alerts useful for public health mitigations at the earliest possible opportunity.
Keywords: Shigella flexneri; multidrug-resistant (MDR); outbreak; surveillance; whole-genome sequence (WGS).
Copyright © 2022 Ko, Chu, Lim, Yingtaweesittikul, Huang, Tan, Goh, Tan, Ng, Maiwald, Chia, Cao, Tan, Sim, Koh, Nagarajan and Suphavilai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Phylogenetic and Evolutionary Analysis Reveals the Recent Dominance of Ciprofloxacin-Resistant Shigella sonnei and Local Persistence of S. flexneri Clones in India.mSphere. 2020 Oct 7;5(5):e00569-20. doi: 10.1128/mSphere.00569-20. mSphere. 2020. PMID: 33028681 Free PMC article.
-
Genome sequence of Shigella flexneri strain SP1, a diarrheal isolate that encodes an extended-spectrum β-lactamase (ESBL).Ann Clin Microbiol Antimicrob. 2017 May 12;16(1):37. doi: 10.1186/s12941-017-0212-2. Ann Clin Microbiol Antimicrob. 2017. PMID: 28499446 Free PMC article.
-
Prevalence and antimicrobial resistance of Shigella flexneri serotype 2 variant in China.Front Microbiol. 2015 May 7;6:435. doi: 10.3389/fmicb.2015.00435. eCollection 2015. Front Microbiol. 2015. PMID: 25999941 Free PMC article.
-
An Outbreak of Multidrug-Resistant Shigella flexneri Serotype 2a Among People Experiencing Homelessness in Vancouver.Trop Med Infect Dis. 2025 Apr 28;10(5):120. doi: 10.3390/tropicalmed10050120. Trop Med Infect Dis. 2025. PMID: 40423350 Free PMC article. Review.
-
Newly Emerged Serotype 1c of Shigella flexneri: Multiple Origins and Changing Drug Resistance Landscape.Genes (Basel). 2020 Sep 3;11(9):1042. doi: 10.3390/genes11091042. Genes (Basel). 2020. PMID: 32899396 Free PMC article.
Cited by
-
Antibiotic-Prescribing Practices for Management of Childhood Diarrhea in 3 Sub-Saharan African Countries: Findings From the Vaccine Impact on Diarrhea in Africa (VIDA) Study, 2015-2018.Clin Infect Dis. 2023 Apr 19;76(76 Suppl1):S32-S40. doi: 10.1093/cid/ciac980. Clin Infect Dis. 2023. PMID: 37074427 Free PMC article.
References
-
- Anderson JD., 4th, Bagamian KH, Muhib F, Amaya MP, Laytner LA, Wierzba T, Rheingans R. Burden of enterotoxigenic Escherichia coli and Shigella non-fatal diarrhoeal infections in 79 low-income and lower middle-income countries: a modelling analysis. Lancet Glob Health. (2019) 7:e321–30. 10.1016/S2214-109X(18)30483-2 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

