Alveolar macrophages in early stage COPD show functional deviations with properties of impaired immune activation
- PMID: 35979364
- PMCID: PMC9377018
- DOI: 10.3389/fimmu.2022.917232
Alveolar macrophages in early stage COPD show functional deviations with properties of impaired immune activation
Abstract
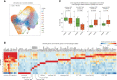
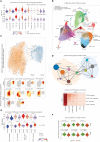
Despite its high prevalence, the cellular and molecular mechanisms of chronic obstructive pulmonary disease (COPD) are far from being understood. Here, we determine disease-related changes in cellular and molecular compositions within the alveolar space and peripheral blood of a cohort of COPD patients and controls. Myeloid cells were the largest cellular compartment in the alveolar space with invading monocytes and proliferating macrophages elevated in COPD. Modeling cell-to-cell communication, signaling pathway usage, and transcription factor binding predicts TGF-β1 to be a major upstream regulator of transcriptional changes in alveolar macrophages of COPD patients. Functionally, macrophages in COPD showed reduced antigen presentation capacity, accumulation of cholesteryl ester, reduced cellular chemotaxis, and mitochondrial dysfunction, reminiscent of impaired immune activation.
Keywords: TGF-β1; blood; bronchoalveolar lavage; chronic obstructive pulmonary disease; impaired immune activation; macrophage; monocyte.
Copyright © 2022 Baßler, Fujii, Kapellos, Dudkin, Reusch, Horne, Reiz, Luecken, Osei-Sarpong, Warnat-Herresthal, Bonaguro, Schulte-Schrepping, Wagner, Günther, Pizarro, Schreiber, Knoll, Holsten, Kröger, De Domenico, Becker, Händler, Wohnhaas, Baumgartner, Köhler, Theis, Kraut, Wadsworth, Hughes, Ferreira, Hinkley, Kaltheuner, Geyer, Thiele, Shalek, Feißt, Thomas, Dickten, Beyer, Baum, Yosef, Aschenbrenner, Ulas, Hasenauer, Theis, Skowasch and Schultze.
Conflict of interest statement
The handling editor [AH] declared a shared affiliation with the author(s) [AW, NY] at the time of review. BR, FB, MK and HD were employed by CommaSoft. CW and PB were employed by Boehringer Ingelheim. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Figures





References
-
- GBD 2017 Causes of Death Collaborators . Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: A systematic analysis for the global burden of disease study 2017. Lancet (2018) 392:1736–88. doi: 10.1016/S0140-6736(18)32203-7 - DOI - PMC - PubMed

