A bioinformatic-assisted workflow for genome-wide identification of ncRNAs
- PMID: 35979446
- PMCID: PMC9376865
- DOI: 10.1093/nargab/lqac059
A bioinformatic-assisted workflow for genome-wide identification of ncRNAs
Abstract
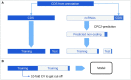
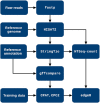
With the upcoming of affordable Next-Generation Sequencing technologies, the number of known non-protein coding RNAs increased drastically in recent years. Different types of non-coding RNAs (ncRNAs) emerged as key players in the regulation of gene expression on the RNA-RNA, RNA-DNA as well as RNA-protein level, ranging from involvement in chromatin remodeling and transcription regulation to post-transcriptional modifications. Prediction of ncRNAs involves the use of several bioinformatics tools and can be a daunting task for researchers. This led to the development of analysis pipelines such as UClncR and lncpipe. However, these pipelines are limited to datasets from human, mouse, zebrafish or fruit fly and are not able to analyze RNA sequencing data from other organisms. In this study, we developed the analysis pipeline Pinc (Pipeline for prediction of ncRNA) as an enhanced tool to predict ncRNAs based on sequencing data by removing transcripts that show protein-coding potential. Additionally, a feature for differential expression analysis of annotated genes as well as for identification of novel ncRNAs is implemented. Pinc uses Nextflow as a framework and is built with robust and well-established analysis tools. This will allow researchers to utilize sequencing data from every organism in order to reliably identify ncRNAs.
© The Author(s) 2022. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures

Similar articles
-
PINC: A Tool for Non-Coding RNA Identification in Plants Based on an Automated Machine Learning Framework.Int J Mol Sci. 2022 Oct 5;23(19):11825. doi: 10.3390/ijms231911825. Int J Mol Sci. 2022. PMID: 36233123 Free PMC article.
-
Detection of RNA structures in porcine EST data and related mammals.BMC Genomics. 2007 Sep 10;8:316. doi: 10.1186/1471-2164-8-316. BMC Genomics. 2007. PMID: 17845718 Free PMC article.
-
nocoRNAc: characterization of non-coding RNAs in prokaryotes.BMC Bioinformatics. 2011 Jan 31;12:40. doi: 10.1186/1471-2105-12-40. BMC Bioinformatics. 2011. PMID: 21281482 Free PMC article.
-
Biogenesis, Functions, Interactions, and Resources of Non-Coding RNAs in Plants.Int J Mol Sci. 2022 Mar 28;23(7):3695. doi: 10.3390/ijms23073695. Int J Mol Sci. 2022. PMID: 35409060 Free PMC article. Review.
-
[Non-coding RNA - from useless to essential].Cas Lek Cesk. 2016 Winter;155(7):370-376. Cas Lek Cesk. 2016. PMID: 27990832 Review. Czech.
Cited by
-
AI-powered precision medicine: utilizing genetic risk factor optimization to revolutionize healthcare.NAR Genom Bioinform. 2025 May 5;7(2):lqaf038. doi: 10.1093/nargab/lqaf038. eCollection 2025 Jun. NAR Genom Bioinform. 2025. PMID: 40330081 Free PMC article. Review.
-
The expression landscape and pangenome of long non-coding RNA in the fungal wheat pathogen Zymoseptoria tritici.Microb Genom. 2023 Nov;9(11):001136. doi: 10.1099/mgen.0.001136. Microb Genom. 2023. PMID: 37991492 Free PMC article.
References
-
- Zhao Q., Sun Y., Wang D., Zhang H., Yu K., Zheng J., Zuo Z.. LncPipe: a Nextflow-based pipeline for identification and analysis of long non-coding RNAs from RNA-Seq data. J. Genet. Genomics. 2018; 45:399–401. - PubMed
LinkOut - more resources
Full Text Sources

