CAPRIN1 haploinsufficiency causes a neurodevelopmental disorder with language impairment, ADHD and ASD
- PMID: 35979925
- PMCID: PMC10169411
- DOI: 10.1093/brain/awac278
CAPRIN1 haploinsufficiency causes a neurodevelopmental disorder with language impairment, ADHD and ASD
Abstract
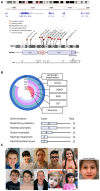
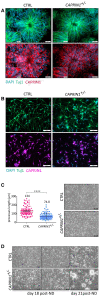
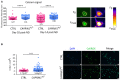
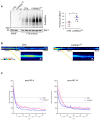
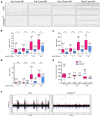
We describe an autosomal dominant disorder associated with loss-of-function variants in the Cell cycle associated protein 1 (CAPRIN1; MIM*601178). CAPRIN1 encodes a ubiquitous protein that regulates the transport and translation of neuronal mRNAs critical for synaptic plasticity, as well as mRNAs encoding proteins important for cell proliferation and migration in multiple cell types. We identified 12 cases with loss-of-function CAPRIN1 variants, and a neurodevelopmental phenotype characterized by language impairment/speech delay (100%), intellectual disability (83%), attention deficit hyperactivity disorder (82%) and autism spectrum disorder (67%). Affected individuals also had respiratory problems (50%), limb/skeletal anomalies (50%), developmental delay (42%) feeding difficulties (33%), seizures (33%) and ophthalmologic problems (33%). In patient-derived lymphoblasts and fibroblasts, we showed a monoallelic expression of the wild-type allele, and a reduction of the transcript and protein compatible with a half dose. To further study pathogenic mechanisms, we generated sCAPRIN1+/- human induced pluripotent stem cells via CRISPR-Cas9 mutagenesis and differentiated them into neuronal progenitor cells and cortical neurons. CAPRIN1 loss caused reduced neuronal processes, overall disruption of the neuronal organization and an increased neuronal degeneration. We also observed an alteration of mRNA translation in CAPRIN1+/- neurons, compatible with its suggested function as translational inhibitor. CAPRIN1+/- neurons also showed an impaired calcium signalling and increased oxidative stress, two mechanisms that may directly affect neuronal networks development, maintenance and function. According to what was previously observed in the mouse model, measurements of activity in CAPRIN1+/- neurons via micro-electrode arrays indicated lower spike rates and bursts, with an overall reduced activity. In conclusion, we demonstrate that CAPRIN1 haploinsufficiency causes a novel autosomal dominant neurodevelopmental disorder and identify morphological and functional alterations associated with this disorder in human neuronal models.
Keywords: ADHD; ASD; CAPRIN1; RNG105; neurodevelopment.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
S.W.S. is on the Scientific Advisory Committees of Population Bio and Deep Genomics, serves as a Highly Cited Academic Advisor for the King Abdulaziz University, and intellectual property from aspects of his research held at the Hospital for Sick Children are licensed to Athena Diagnostics and Population Bio.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

