Biofilm Formation in Streptococcus agalactiae Is Inhibited by a Small Regulatory RNA Regulated by the Two-Component System CiaRH
- PMID: 35980045
- PMCID: PMC9603419
- DOI: 10.1128/spectrum.00635-22
Biofilm Formation in Streptococcus agalactiae Is Inhibited by a Small Regulatory RNA Regulated by the Two-Component System CiaRH
Abstract
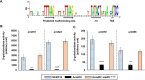
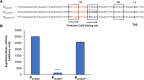
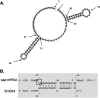
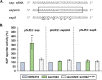
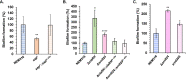
Regulatory small RNAs (sRNAs) are involved in the adaptation of bacteria to their environment. CiaR-dependent sRNAs (csRNAs) are controlled by the regulatory two-component system (TCS) CiaRH, which is widely conserved in streptococci. Except for Streptococcus pneumoniae and Streptococcus sanguinis, the targets of these csRNAs have not yet been investigated. Streptococcus agalactiae, the leading cause of neonatal infections, has four conserved csRNA genes, namely, srn015, srn024, srn070, and srn085. Here, we demonstrate the importance of the direct repeat TTTAAG-N5-TTTAAG in the regulation of these csRNAs by CiaRH. A 24-nucleotide Srn024-sap RNA base-pairing region is predicted in silico. The sap gene encodes a LPXTG-cell wall-anchored pullulanase. This protein cleaves α-glucan polysaccharides such as pullulan and glycogen present in the environment to release glucose and is involved in adhesion to human cervical epithelial cells. Inactivation of S. agalactiae pullulanase (SAP) leads to no bacterial growth in a medium with only pullulan as a carbon source and reduced biofilm formation, while deletion of ciaRH and srn024 genes significantly increases bacterial growth and biofilm formation. Using a new translational fusion vector, we demonstrated that Srn024 is involved in the posttranscriptional regulation of sap expression. Complementary base pair exchanges in S. agalactiae suggest that Srn024 interacts directly with sap mRNA and that disruption of this RNA pairing is sufficient to yield the biofilm phenotype of Srn024 deletion. These results suggest the involvement of Srn024 in the adaptation of S. agalactiae to environmental changes and biofilm formation, likely through the regulation of the sap gene. IMPORTANCE Although Streptococcus agalactiae is a commensal bacterium of the human digestive and genitourinary tracts, it is also an opportunistic pathogen for humans and other animals. As the main cause of neonatal infections, it is responsible for pneumonia, bacteremia, and meningitis. However, its adaptation to these different ecological niches is not fully understood. Bacterial regulatory networks are involved in this adaptation, and the regulatory TCSs (e.g., CiaRH), as well as the regulatory sRNAs, are part of it. This study is the first step to understand the role of csRNAs in the adaptation of S. agalactiae. This bacterium does not currently exhibit extensive antibiotic resistance. However, it is crucial to find alternatives before multidrug resistance emerges. Therefore, we propose that drugs targeting regulatory RNAs with Srn024-like activities would affect pathogens by reducing their abilities to form biofilm and to adapt to host niches.
Keywords: CiaRH; SAP pullulanase; Streptococcus agalactiae; adaptation; biofilm; csRNAs; regulation; sRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Evans JJ, Bohnsack JF, Klesius PH, Whiting AA, Garcia JC, Shoemaker CA, Takahashi S. 2008. Phylogenetic relationships among Streptococcus agalactiae isolated from piscine, dolphin, bovine and human sources: a dolphin and piscine lineage associated with a fish epidemic in Kuwait is also associated with human neonatal infections in Japan. J Med Microbiol 57:1369–1376. doi:10.1099/jmm.0.47815-0. - DOI - PubMed
-
- van der Mee-Marquet N, Fourny L, Arnault L, Domelier A-S, Salloum M, Lartigue M-F, Quentin R. 2008. Molecular characterization of human-colonizing Streptococcus agalactiae strains isolated from throat, skin, anal margin, and genital body sites. J Clin Microbiol 46:2906–2911. doi:10.1128/JCM.00421-08. - DOI - PMC - PubMed
-
- Stoll BJ, Hansen NI, Sánchez PJ, Faix RG, Poindexter BB, Van Meurs KP, Bizzarro MJ, Goldberg RN, Frantz ID, Hale EC, Shankaran S, Kennedy K, Carlo WA, Watterberg KL, Bell EF, Walsh MC, Schibler K, Laptook AR, Shane AL, Schrag SJ, Das A, Higgins RD. 2011. Early onset neonatal sepsis: the burden of group B streptococcal and E. coli disease continues. Pediatrics 127:817–826. doi:10.1542/peds.2010-2217. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

