Monitoring SARS-CoV-2 Infection Using a Double Reporter-Expressing Virus
- PMID: 35980204
- PMCID: PMC9603146
- DOI: 10.1128/spectrum.02379-22
Monitoring SARS-CoV-2 Infection Using a Double Reporter-Expressing Virus
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the highly contagious agent responsible for the coronavirus disease 2019 (COVID-19) pandemic. An essential requirement for understanding SARS-CoV-2 biology and the impact of antiviral therapeutics is a robust method to detect the presence of the virus in infected cells or animal models. Despite the development and successful generation of recombinant (r)SARS-CoV-2-expressing fluorescent or luciferase reporter genes, knowledge acquired from their use in in vitro assays and/or in live animals is limited to the properties of the fluorescent or luciferase reporter genes. Herein, for the first time, we engineered a replication-competent rSARS-CoV-2 that expresses both fluorescent (mCherry) and luciferase (Nluc) reporter genes (rSARS-CoV-2/mCherry-Nluc) to overcome limitations associated with the use of a single reporter gene. In cultured cells, rSARS-CoV-2/mCherry-Nluc displayed similar viral fitness as rSARS-CoV-2 expressing single reporter fluorescent and luciferase genes (rSARS-CoV-2/mCherry and rSARS-CoV-2/Nluc, respectively) or wild-type (WT) rSARS-CoV-2, while maintaining comparable expression levels of both reporter genes. In vivo, rSARS-CoV-2/mCherry-Nluc has similar pathogenicity in K18 human angiotensin-converting enzyme 2 (hACE2) transgenic mice than rSARS-CoV-2 expressing individual reporter genes or WT rSARS-CoV-2. Importantly, rSARS-CoV-2/mCherry-Nluc facilitates the assessment of viral infection and transmission in golden Syrian hamsters using in vivo imaging systems (IVIS). Altogether, this study demonstrates the feasibility of using this novel bioreporter-expressing rSARS-CoV-2 for the study of SARS-CoV-2 in vitro and in vivo. IMPORTANCE Despite the availability of vaccines and antivirals, the coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to ravage health care institutions worldwide. Previously, we generated replication-competent recombinant (r)SARS-CoV-2 expressing fluorescent or luciferase reporter proteins to track viral infection in vitro and/or in vivo. However, these rSARS-CoV-2 are restricted to express only a single fluorescent or a luciferase reporter gene, limiting or preventing their use in specific in vitro assays and/or in vivo studies. To overcome this limitation, we have engineered a rSARS-CoV-2 expressing both fluorescent (mCherry) and luciferase (Nluc) genes and demonstrated its feasibility to study the biology of SARS-CoV-2 in vitro and/or in vivo, including the identification and characterization of neutralizing antibodies and/or antivirals. Using rodent models, we visualized SARS-CoV-2 infection and transmission through in vivo imaging systems (IVIS).
Keywords: SARS-CoV-2; coronavirus; fluorescent; infection; luciferase; recombinant; reporter; transmission.
Conflict of interest statement
The authors declare a conflict of interest. C.Y. and L.M.-S. are co-inventors on a patent that includes claims related to reverse genetics approaches to generate recombinant SARS-CoV-2.
Figures

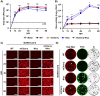


Similar articles
-
Live Imaging and Quantification of Viral Infection in K18 hACE2 Transgenic Mice Using Reporter-Expressing Recombinant SARS-CoV-2.J Vis Exp. 2021 Nov 5;(177):10.3791/63127. doi: 10.3791/63127. J Vis Exp. 2021. PMID: 34806704 Free PMC article.
-
Generation and Characterization of recombinant SARS-CoV-2 expressing reporter genes.J Virol. 2021 Mar 10;95(7):e02209-20. doi: 10.1128/JVI.02209-20. Epub 2021 Jan 11. J Virol. 2021. PMID: 33431557 Free PMC article.
-
A Bifluorescent-Based Assay for the Identification of Neutralizing Antibodies against SARS-CoV-2 Variants of Concern In Vitro and In Vivo.J Virol. 2021 Oct 27;95(22):e0112621. doi: 10.1128/JVI.01126-21. Epub 2021 Sep 8. J Virol. 2021. PMID: 34495697 Free PMC article.
-
K18- and CAG-hACE2 Transgenic Mouse Models and SARS-CoV-2: Implications for Neurodegeneration Research.Molecules. 2022 Jun 28;27(13):4142. doi: 10.3390/molecules27134142. Molecules. 2022. PMID: 35807384 Free PMC article. Review.
-
Rapid Development of Small Rodent Animal Models for Infectious Disease Research Through Vectorized Receptor Molecule Expression.Viruses. 2024 Nov 19;16(11):1794. doi: 10.3390/v16111794. Viruses. 2024. PMID: 39599908 Free PMC article. Review.
Cited by
-
A novel high-throughput screen identifies phenazine-1-carboxylic acid as an inhibitor of African swine fever virus replication in primary porcine alveolar macrophages.Vet Res. 2025 Feb 8;56(1):37. doi: 10.1186/s13567-025-01467-2. Vet Res. 2025. PMID: 39923101 Free PMC article.
-
Development and characterization of a recombinant Senecavirus A expressing enhanced green fluorescent protein.Front Microbiol. 2024 Sep 26;15:1443696. doi: 10.3389/fmicb.2024.1443696. eCollection 2024. Front Microbiol. 2024. PMID: 39391602 Free PMC article.
-
Novel replication-competent reporter-expressing Rift Valley fever viruses for molecular studies.J Virol. 2025 Jan 31;99(1):e0178224. doi: 10.1128/jvi.01782-24. Epub 2024 Dec 12. J Virol. 2025. PMID: 39665546 Free PMC article.
-
Development and validation of Mayaro virus with luciferase reporter genes as a tool for antiviral assays.Heliyon. 2024 Jul 2;10(13):e33885. doi: 10.1016/j.heliyon.2024.e33885. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39071632 Free PMC article.
-
Fluorescent and Bioluminescent Reporter Mouse-Adapted Ebola Viruses Maintain Pathogenicity and Can Be Visualized in Vivo.J Infect Dis. 2023 Nov 15;228(Suppl 7):S536-S547. doi: 10.1093/infdis/jiad136. J Infect Dis. 2023. PMID: 37145895 Free PMC article.
References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi:10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Gudbjartsson DF, Helgason A, Jonsson H, Magnusson OT, Melsted P, Norddahl GL, Saemundsdottir J, Sigurdsson A, Sulem P, Agustsdottir AB, Eiriksdottir B, Fridriksdottir R, Gardarsdottir EE, Georgsson G, Gretarsdottir OS, Gudmundsson KR, Gunnarsdottir TR, Gylfason A, Holm H, Jensson BO, Jonasdottir A, Jonsson F, Josefsdottir KS, Kristjansson T, Magnusdottir DN, le Roux L, Sigmundsdottir G, Sveinbjornsson G, Sveinsdottir KE, Sveinsdottir M, Thorarensen EA, Thorbjornsson B, Löve A, Masson G, Jonsdottir I, Möller AD, Gudnason T, Kristinsson KG, Thorsteinsdottir U, Stefansson K. 2020. Spread of SARS-CoV-2 in the Icelandic population. N Engl J Med 382:2302–2315. doi:10.1056/NEJMoa2006100. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

