UBR5 Acts as an Antiviral Host Factor against MERS-CoV via Promoting Ubiquitination and Degradation of ORF4b
- PMID: 35980206
- PMCID: PMC9472757
- DOI: 10.1128/jvi.00741-22
UBR5 Acts as an Antiviral Host Factor against MERS-CoV via Promoting Ubiquitination and Degradation of ORF4b
Abstract
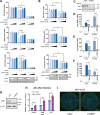
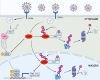
Within the past 2 decades, three highly pathogenic human coronaviruses have emerged, namely, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The health threats and economic burden posed by these tremendously severe coronaviruses have paved the way for research on their etiology, pathogenesis, and treatment. Compared to SARS-CoV and SARS-CoV-2, MERS-CoV genome encoded fewer accessory proteins, among which the ORF4b protein had anti-immunity ability in both the cytoplasm and nucleus. Our work for the first time revealed that ORF4b protein was unstable in the host cells and could be degraded by the ubiquitin proteasome system. After extensive screenings, it was found that UBR5 (ubiquitin protein ligase E3 component N-recognin 5), a member of the HECT E3 ubiquitin ligases, specifically regulated the ubiquitination and degradation of ORF4b. Similar to ORF4b, UBR5 can also translocate into the nucleus through its nuclear localization signal, enabling it to regulate ORF4b stability in both the cytoplasm and nucleus. Through further experiments, lysine 36 was identified as the ubiquitination site on the ORF4b protein, and this residue was highly conserved in various MERS-CoV strains isolated from different regions. When UBR5 was knocked down, the ability of ORF4b to suppress innate immunity was enhanced and MERS-CoV replication was stronger. As an anti-MERS-CoV host protein, UBR5 targets and degrades ORF4b protein through the ubiquitin proteasome system, thereby attenuating the anti-immunity ability of ORF4b and ultimately inhibiting MERS-CoV immune escape, which is a novel antagonistic mechanism of the host against MERS-CoV infection. IMPORTANCE ORF4b was an accessory protein unique to MERS-CoV and was not present in SARS-CoV and SARS-CoV-2 which can also cause severe respiratory disease. Moreover, ORF4b inhibited the production of antiviral cytokines in both the cytoplasm and the nucleus, which was likely to be associated with the high lethality of MERS-CoV. However, whether the host proteins regulate the function of ORF4b is unknown. Our study first determined that UBR5, a host E3 ligase, was a potential host anti-MERS-CoV protein that could reduce the protein level of ORF4b and diminish its anti-immunity ability by inducing ubiquitination and degradation. Based on the discovery of ORF4b-UBR5, a critical molecular target, further increasing the degradation of ORF4b caused by UBR5 could provide a new strategy for the clinical development of drugs for MERS-CoV.
Keywords: IRF3; MERS-CoV; accessory protein; antiviral mechanism; coronavirus; posttranslational modification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Multiple E3 ligases act as antiviral factors against SARS-CoV-2 via inducing the ubiquitination and degradation of ORF9b.J Virol. 2024 Jun 13;98(6):e0162423. doi: 10.1128/jvi.01624-23. Epub 2024 May 6. J Virol. 2024. PMID: 38709105 Free PMC article.
-
Host E3 ligase HUWE1 attenuates the proapoptotic activity of the MERS-CoV accessory protein ORF3 by promoting its ubiquitin-dependent degradation.J Biol Chem. 2022 Feb;298(2):101584. doi: 10.1016/j.jbc.2022.101584. Epub 2022 Jan 13. J Biol Chem. 2022. PMID: 35032548 Free PMC article.
-
Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets.Sci Rep. 2015 Dec 3;5:17554. doi: 10.1038/srep17554. Sci Rep. 2015. PMID: 26631542 Free PMC article.
-
The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds.Antiviral Res. 2015 Mar;115:21-38. doi: 10.1016/j.antiviral.2014.12.015. Epub 2014 Dec 29. Antiviral Res. 2015. PMID: 25554382 Free PMC article. Review.
-
A comparative review of pathogenesis and host innate immunity evasion strategies among the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV).Arch Microbiol. 2021 Jul;203(5):1943-1951. doi: 10.1007/s00203-021-02265-y. Epub 2021 Mar 7. Arch Microbiol. 2021. PMID: 33682075 Free PMC article. Review.
Cited by
-
Multiple E3 ligases act as antiviral factors against SARS-CoV-2 via inducing the ubiquitination and degradation of ORF9b.J Virol. 2024 Jun 13;98(6):e0162423. doi: 10.1128/jvi.01624-23. Epub 2024 May 6. J Virol. 2024. PMID: 38709105 Free PMC article.
-
Lung-Targeted Lipid Nanoparticle-Delivered siUSP33 Attenuates SARS-CoV-2 Replication and Virulence by Promoting Envelope Degradation.Adv Sci (Weinh). 2024 Nov;11(42):e2406211. doi: 10.1002/advs.202406211. Epub 2024 Sep 20. Adv Sci (Weinh). 2024. PMID: 39301916 Free PMC article.
-
A review: targeting UBR5 domains to mediate emerging roles and mechanisms - chance or necessity?Int J Surg. 2024 Aug 1;110(8):4947-4964. doi: 10.1097/JS9.0000000000001541. Int J Surg. 2024. PMID: 38701508 Free PMC article. Review.
-
Extended insights into the pathophysiological role of UBR5: a commentary.Int J Surg. 2024 Sep 1;110(9):6024-6025. doi: 10.1097/JS9.0000000000001716. Int J Surg. 2024. PMID: 39275785 Free PMC article. No abstract available.
-
E3 ubiquitin ligase UBR5 modulates circadian rhythm by facilitating the ubiquitination and degradation of the key clock transcription factor BMAL1.Acta Pharmacol Sin. 2024 Sep;45(9):1793-1808. doi: 10.1038/s41401-024-01290-z. Epub 2024 May 13. Acta Pharmacol Sin. 2024. PMID: 38740904
References
-
- Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. 2020. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27:325–328. 10.1016/j.chom.2020.02.001. - DOI - PMC - PubMed
-
- Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, Flemban H, Al-Nassir WN, Balkhy HH, Al-Hakeem RF, Makhdoom HQ, Zumla AI, Memish ZA. 2013. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 13:752–761. 10.1016/S1473-3099(13)70204-4. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

