TGF-β regulates the stem-like state of PD-1+ TCF-1+ virus-specific CD8 T cells during chronic infection
- PMID: 35980386
- PMCID: PMC9393409
- DOI: 10.1084/jem.20211574
TGF-β regulates the stem-like state of PD-1+ TCF-1+ virus-specific CD8 T cells during chronic infection
Abstract
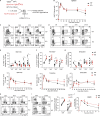
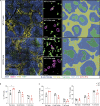
Recent studies have defined a novel population of PD-1+ TCF-1+ stem-like CD8 T cells in chronic infections and cancer. These quiescent cells reside in lymphoid tissues, are critical for maintaining the CD8 T cell response under conditions of persistent antigen, and provide the proliferative burst after PD-1 blockade. Here we examined the role of TGF-β in regulating the differentiation of virus-specific CD8 T cells during chronic LCMV infection of mice. We found that TGF-β signaling was not essential for the generation of the stem-like CD8 T cells but was critical for maintaining the stem-like state and quiescence of these cells. TGF-β regulated the unique transcriptional program of the stem-like subset, including upregulation of inhibitory receptors specifically expressed on these cells. TGF-β also promoted the terminal differentiation of exhausted CD8 T cells by suppressing the effector-associated program. Together, the absence of TGF-β signaling resulted in significantly increased accumulation of effector-like CD8 T cells. These findings have implications for immunotherapies in general and especially for T cell therapy against chronic infections and cancer.
© 2022 Hu et al.
Conflict of interest statement
Disclosures: Y. Hu reported a patent for inhibitory molecules specifically expressed by a subset of stem-like CD8 T cells for therapeutic intervention pending. C. Medina reported a patent for inhibitory molecules specifically expressed by a subset of stem-like CD8 T cells for therapeutic intervention pending. S.J. Turley is an employee of Genentech/Roche. R. Ahmed reported a patent for monoclonal antibodies directed against PD-1 issued and a patent for inhibitory molecules specifically expressed by a subset of stem-like CD8 T cells for therapeutic intervention pending. No other disclosures were reported.
Figures













References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

