Chemical zymogens for the protein cysteinome
- PMID: 35982075
- PMCID: PMC9388531
- DOI: 10.1038/s41467-022-32609-1
Chemical zymogens for the protein cysteinome
Abstract
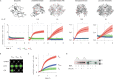
We present three classes of chemical zymogens established around the protein cysteinome. In each case, the cysteine thiol group was converted into a mixed disulfide: with a small molecule, a non-degradable polymer, or with a fast-depolymerizing fuse polymer (ZLA). The latter was a polydisulfide based on naturally occurring molecule, lipoic acid. Zymogen designs were applied to cysteine proteases and a kinase. In each case, enzymatic activity was successfully masked in full and reactivated by small molecule reducing agents. However, only ZLA could be reactivated by protein activators, demonstrating that the macromolecular fuse escapes the steric bulk created by the protein globule, collects activation signal in solution, and relays it to the active site of the enzyme. This afforded first-in-class chemical zymogens that are activated via protein-protein interactions. We also document zymogen exchange reactions whereby the polydisulfide is transferred between the interacting proteins via the "chain transfer" bioconjugation mechanism.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

