Computational simulation of liver fibrosis dynamics
- PMID: 35982187
- PMCID: PMC9388486
- DOI: 10.1038/s41598-022-18123-w
Computational simulation of liver fibrosis dynamics
Abstract
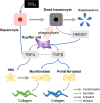
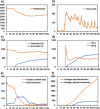
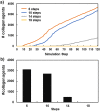
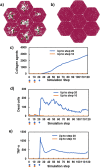
Liver fibrosis is a result of homeostasis breakdown caused by repetitive injury. The accumulation of collagens disrupts liver structure and function, which causes serious consequences such as cirrhosis. Various mathematical simulation models have been developed to understand these complex processes. We employed the agent-based modelling (ABM) approach and implemented inflammatory processes in central venous regions. Collagens were individually modelled and visualised depending on their origin: myofibroblast and portal fibroblast. Our simulation showed that the administration of toxic compounds induced accumulation of myofibroblast-derived collagens in central venous regions and portal fibroblast-derived collagens in portal areas. Subsequently, these collagens were bridged between central-central areas and spread all over areas. We confirmed the consistent dynamic behaviour of collagen formulation in our simulation and from histological sections obtained via in vivo experiments. Sensitivity analyses identified dead hepatocytes caused by inflammation and the ratio of residential liver cells functioned as a cornerstone for the initiation and progression of liver fibrosis. The validated mathematical model demonstrated here shows virtual experiments that are complementary to biological experiments, which contribute to understanding a new mechanism of liver fibrosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

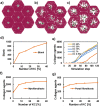
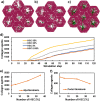
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

