Evidence of a genetically driven metabolomic signature in actively inflamed Crohn's disease
- PMID: 35982195
- PMCID: PMC9388636
- DOI: 10.1038/s41598-022-18178-9
Evidence of a genetically driven metabolomic signature in actively inflamed Crohn's disease
Abstract
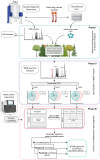
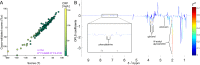
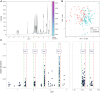
Crohn's disease (CD) is characterised by chronic inflammation. We aimed to identify a relationship between plasma inflammatory metabolomic signature and genomic data in CD using blood plasma metabolic profiles. Proton NMR spectroscopy were achieved for 228 paediatric CD patients. Regression (OPLS) modelling and machine learning (ML) approaches were independently applied to establish the metabolic inflammatory signature, which was correlated against gene-level pathogenicity scores generated for all patients and functional enrichment was analysed. OPLS modelling of metabolomic spectra from unfasted patients revealed distinctive shifts in plasma metabolites corresponding to regions of the spectrum assigned to N-acetyl glycoprotein, glycerol and phenylalanine that were highly correlated (R2 = 0.62) with C-reactive protein levels. The same metabolomic signature was independently identified using ML to predict patient inflammation status. Correlation of the individual peaks comprising this metabolomic signature of inflammation with pathogenic burden across 15,854 unselected genes identified significant enrichment for genes functioning within 'intrinsic component of membrane' (p = 0.003) and 'inflammatory bowel disease (IBD)' (p = 0.003). The seven genes contributing IBD enrichment are critical regulators of pro-inflammatory signaling. Overall, a metabolomic signature of inflammation can be detected from blood plasma in CD. This signal is correlated with pathogenic mutation in pro-inflammatory immune response genes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

