Temporal lineage replacements and dominance of imported variants of concern during the COVID-19 pandemic in Kenya
- PMID: 35982756
- PMCID: PMC9382597
- DOI: 10.1038/s43856-022-00167-8
Temporal lineage replacements and dominance of imported variants of concern during the COVID-19 pandemic in Kenya
Abstract
Background: Kenya's COVID-19 epidemic was seeded early in March 2020 and did not peak until early August 2020 (wave 1), late-November 2020 (wave 2), mid-April 2021 (wave 3), late August 2021 (wave 4), and mid-January 2022 (wave 5).
Methods: Here, we present SARS-CoV-2 lineages associated with the five waves through analysis of 1034 genomes, which included 237 non-variants of concern and 797 variants of concern (VOC) that had increased transmissibility, disease severity or vaccine resistance.
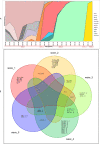
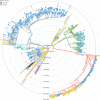
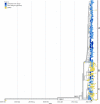
Results: In total 40 lineages were identified. The early European lineages (B.1 and B.1.1) were the first to be seeded. The B.1 lineage continued to expand and remained dominant, accounting for 60% (72/120) and 57% (45/79) in waves 1 and 2 respectively. Waves three, four and five respectively were dominated by VOCs that were distributed as follows: Alpha 58.5% (166/285), Delta 92.4% (327/354), Omicron 95.4% (188/197) and Beta at 4.2% (12/284) during wave 3 and 0.3% (1/354) during wave 4. Phylogenetic analysis suggests multiple introductions of variants from outside Kenya, more so during the first, third, fourth and fifth waves, as well as subsequent lineage diversification.
Conclusions: The data highlights the importance of genome surveillance in determining circulating variants to aid interpretation of phenotypes such as transmissibility, virulence and/or resistance to therapeutics/vaccines.
Keywords: Respiratory distress syndrome; Viral infection.
© The Author(s) 2022.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

