A microengineered Brain-Chip to model neuroinflammation in humans
- PMID: 35982785
- PMCID: PMC9379671
- DOI: 10.1016/j.isci.2022.104813
A microengineered Brain-Chip to model neuroinflammation in humans
Abstract
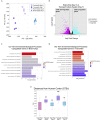
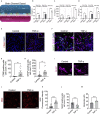
Species differences in brain and blood-brain barrier (BBB) biology hamper the translation of findings from animal models to humans, impeding the development of therapeutics for brain diseases. Here, we present a human organotypic microphysiological system (MPS) that includes endothelial-like cells, pericytes, glia, and cortical neurons and maintains BBB permeability at in vivo relevant levels. This human Brain-Chip engineered to recapitulate critical aspects of the complex interactions that mediate neuroinflammation and demonstrates significant improvements in clinical mimicry compared to previously reported similar MPS. In comparison to Transwell culture, the transcriptomic profiling of the Brain-Chip displayed significantly advanced similarity to the human adult cortex and enrichment in key neurobiological pathways. Exposure to TNF-α recreated the anticipated inflammatory environment shown by glia activation, increased release of proinflammatory cytokines, and compromised barrier permeability. We report the development of a robust brain MPS for mechanistic understanding of cell-cell interactions and BBB function during neuroinflammation.
Keywords: Biomedical engineering; Cellular neuroscience; Molecular neuroscience.
© 2022 The Authors.
Conflict of interest statement
Patents covering all the main aspects of the use of human Brain-Chip as a model to study neuroinflammation have been filed by Emulate, Inc. (application numbers: US16/712,439 and PCT/US2020/056,245). The applications are currently pending. I.P., K.R.K., D.V.M., C.Y.L., S.B., A.S., L.E., C.D.H., and K.K. are current or former employees of Emulate, Inc and may hold equity interests in Emulate, Inc. All other authors declare no competing interests.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

