Digging for the discovery of SARS-CoV-2 nsp12 inhibitors: a pharmacophore-based and molecular dynamics simulation study
- PMID: 35983350
- PMCID: PMC9370102
- DOI: 10.2217/fvl-2022-0054
Digging for the discovery of SARS-CoV-2 nsp12 inhibitors: a pharmacophore-based and molecular dynamics simulation study
Abstract
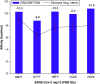
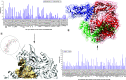
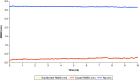
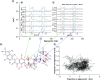
Aim: COVID-19 is a global health threat. Therapeutics are urgently needed to cure patients severely infected with COVID-19. Objective: to investigate potential candidates of nsp12 inhibitors by searching for druggable cavity pockets within the viral protein and drug discovery. Methods: A virtual screening of ZINC natural products on SARS-CoV-2 nsp12's druggable cavity was performed. A lead compound with the highest affinity to nsp12 was simulated dynamically for 10 ns. Results: ZINC03977803 was nominated as the lead compound. The results showed stable interaction between ZINC03977803 and nsp12 during 10 ns. Discussion: ZINC03977803 showed stable interaction with the catalytic subunit of SARS-CoV-2, nsp12. It could inhibit the SARS-CoV-2 life cycle by direct interaction with nsp12 and inhibit RdRp complex formation.
Keywords: RNA-dependent RNA polymerase; SARS-CoV-2; molecular docking; molecular dynamics; natural product; pharmacophore-based drug discovery.
© 2022 Future Medicine Ltd.
Figures





References
-
- Mohebbi A, Askari FS, Ebrahimi M et al. Susceptibility of the Iranian population to severe acute respiratory syndrome coronavirus 2 infection based on variants of angiotensin I converting enzyme 2. Future Virol. 15(8), 507–514 (2020).
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
