Proteasomal Degradation of TRAF2 Mediates Mitochondrial Dysfunction in Doxorubicin-Cardiomyopathy
- PMID: 35983756
- PMCID: PMC10043946
- DOI: 10.1161/CIRCULATIONAHA.121.058411
Proteasomal Degradation of TRAF2 Mediates Mitochondrial Dysfunction in Doxorubicin-Cardiomyopathy
Erratum in
-
Correction to: Proteasomal Degradation of TRAF2 Mediates Mitochondrial Dysfunction in Doxorubicin-Cardiomyopathy.Circulation. 2022 Oct 11;146(15):e224. doi: 10.1161/CIR.0000000000001108. Epub 2022 Oct 10. Circulation. 2022. PMID: 36214137 No abstract available.
Abstract
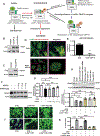
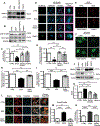
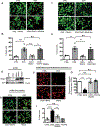
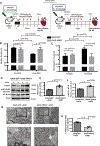
Background: Cytokines such as tumor necrosis factor-α (TNFα) have been implicated in cardiac dysfunction and toxicity associated with doxorubicin (DOX). Although TNFα can elicit different cellular responses, including survival or death, the mechanisms underlying these divergent outcomes in the heart remain cryptic. The E3 ubiquitin ligase TRAF2 (TNF receptor associated factor 2) provides a critical signaling platform for K63-linked polyubiquitination of RIPK1 (receptor interacting protein 1), crucial for nuclear factor-κB (NF-κB) activation by TNFα and survival. Here, we investigate alterations in TNFα-TRAF2-NF-κB signaling in the pathogenesis of DOX cardiotoxicity.
Methods: Using a combination of in vivo (4 weekly injections of DOX 5 mg·kg-1·wk-1) in C57/BL6J mice and in vitro approaches (rat, mouse, and human inducible pluripotent stem cell-derived cardiac myocytes), we monitored TNFα levels, lactate dehydrogenase, cardiac ultrastructure and function, mitochondrial bioenergetics, and cardiac cell viability.
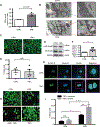
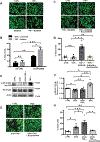
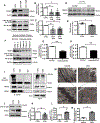
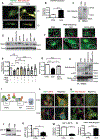
Results: In contrast to vehicle-treated mice, ultrastructural defects, including cytoplasmic swelling, mitochondrial perturbations, and elevated TNFα levels, were observed in the hearts of mice treated with DOX. While investigating the involvement of TNFα in DOX cardiotoxicity, we discovered that NF-κB was readily activated by TNFα. However, TNFα-mediated NF-κB activation was impaired in cardiac myocytes treated with DOX. This coincided with loss of K63- linked polyubiquitination of RIPK1 from the proteasomal degradation of TRAF2. Furthermore, TRAF2 protein abundance was markedly reduced in hearts of patients with cancer treated with DOX. We further established that the reciprocal actions of the ubiquitinating and deubiquitinating enzymes cellular inhibitors of apoptosis 1 and USP19 (ubiquitin-specific peptidase 19), respectively, regulated the proteasomal degradation of TRAF2 in DOX-treated cardiac myocytes. An E3-ligase mutant of cellular inhibitors of apoptosis 1 (H588A) or gain of function of USP19 prevented proteasomal degradation of TRAF2 and DOX-induced cell death. Furthermore, wild-type TRAF2, but not a RING finger mutant defective for K63-linked polyubiquitination of RIPK1, restored NF-κB signaling and suppressed DOX-induced cardiac cell death. Last, cardiomyocyte-restricted expression of TRAF2 (cardiac troponin T-adeno-associated virus 9-TRAF2) in vivo protected against mitochondrial defects and cardiac dysfunction induced by DOX.
Conclusions: Our findings reveal a novel signaling axis that functionally connects the cardiotoxic effects of DOX to proteasomal degradation of TRAF2. Disruption of the critical TRAF2 survival pathway by DOX sensitizes cardiac myocytes to TNFα-mediated necrotic cell death and DOX cardiotoxicity.
Keywords: TNF receptor-associated factor 2; doxorubicin; inhibitor of apoptosis proteins; mitochondria; myocytes, cardiac; proteasome endopeptidase complex; tumor necrosis factor-alpha.
Conflict of interest statement
Competing interests
No competing interests.
Figures

Comment in
-
Response by Dhingra et al to Letter Regarding Article, "Proteasomal Degradation of TRAF2 Mediates Mitochondrial Dysfunction in Doxorubicin-Cardiomyopathy".Circulation. 2023 Mar 28;147(13):1051-1052. doi: 10.1161/CIRCULATIONAHA.123.063546. Epub 2023 Mar 27. Circulation. 2023. PMID: 36972342 Free PMC article. No abstract available.
-
Letter by Shi et al Regarding Article, "Proteasomal Degradation of TRAF2 Mediates Mitochondrial Dysfunction in Doxorubicin-Cardiomyopathy".Circulation. 2023 Mar 28;147(13):1050. doi: 10.1161/CIRCULATIONAHA.122.063010. Epub 2023 Mar 27. Circulation. 2023. PMID: 36972345 No abstract available.
References
-
- Dhingra R, Margulets V, Chowdhury SR, Thliveris J, Jassal D, Fernyhough P, Dorn GW, Kirshenbaum LA. Bnip3 mediates doxorubicin-induced cardiac myocyte necrosis and mortality through changes in mitochondrial signaling. Proc Natl Acad Sci. 2014;111:E5537–E5544. doi: 10.1073/pnas.1414665111. - DOI - PMC - PubMed
-
- Bradley JR. TNF-mediated inflammatory disease. J. Pathol. 2008;214:149–160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

