Identification of growth regulators using cross-species network analysis in plants
- PMID: 35984294
- PMCID: PMC9706488
- DOI: 10.1093/plphys/kiac374
Identification of growth regulators using cross-species network analysis in plants
Abstract
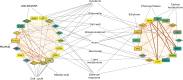
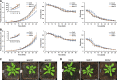
With the need to increase plant productivity, one of the challenges plant scientists are facing is to identify genes that play a role in beneficial plant traits. Moreover, even when such genes are found, it is generally not trivial to transfer this knowledge about gene function across species to identify functional orthologs. Here, we focused on the leaf to study plant growth. First, we built leaf growth transcriptional networks in Arabidopsis (Arabidopsis thaliana), maize (Zea mays), and aspen (Populus tremula). Next, known growth regulators, here defined as genes that when mutated or ectopically expressed alter plant growth, together with cross-species conserved networks, were used as guides to predict novel Arabidopsis growth regulators. Using an in-depth literature screening, 34 out of 100 top predicted growth regulators were confirmed to affect leaf phenotype when mutated or overexpressed and thus represent novel potential growth regulators. Globally, these growth regulators were involved in cell cycle, plant defense responses, gibberellin, auxin, and brassinosteroid signaling. Phenotypic characterization of loss-of-function lines confirmed two predicted growth regulators to be involved in leaf growth (NPF6.4 and LATE MERISTEM IDENTITY2). In conclusion, the presented network approach offers an integrative cross-species strategy to identify genes involved in plant growth and development.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, Lenhard M (2007) Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell 13: 843–856 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

