BluePrint breast cancer molecular subtyping recognizes single and dual subtype tumors with implications for therapeutic guidance
- PMID: 35984580
- PMCID: PMC9464757
- DOI: 10.1007/s10549-022-06698-x
BluePrint breast cancer molecular subtyping recognizes single and dual subtype tumors with implications for therapeutic guidance
Abstract
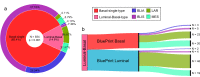
Purpose: BluePrint (BP) is an 80-gene molecular subtyping test that classifies early-stage breast cancer (EBC) into Basal, Luminal, and HER2 subtypes. In most cases, breast tumors have one dominant subtype, representative of a single activated pathway. However, some tumors show a statistically equal representation of more than one subtype, referred to as dual subtype. This study aims to identify and examine dual subtype tumors by BP to understand their biology and possible implications for treatment guidance.
Methods: The BP scores of over 15,000 tumor samples from EBC patients were analyzed, and the differences between the highest and the lowest scoring subtypes were calculated. Based upon the distribution of the differences between BP scores, a threshold was determined for each subtype to identify dual versus single subtypes.
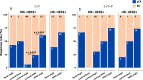
Results: Approximately 97% of samples had one single activated BluePrint molecular subtype, whereas ~ 3% of samples were classified as BP dual subtype. The most frequently occurring dual subtypes were the Luminal-Basal-type and Luminal-HER2-type. Luminal-Basal-type displays a distinct biology from the Luminal single type and Basal single type. Burstein's classification of the single and dual Basal samples showed that the Luminal-Basal-type is mostly classified as 'luminal androgen receptor' and 'mesenchymal' subtypes, supporting molecular evidence of AR activation in the Luminal-Basal-type tumors. Tumors classified as Luminal-HER2-type resemble features of both Luminal-single-type and HER2-single-type. However, patients with dual Luminal-HER2-type have a lower pathological complete response after receiving HER2-targeted therapies in addition to chemotherapy in comparison with patients with a HER2-single-type.
Conclusion: This study demonstrates that BP identifies tumors with two active functional pathways (dual subtype) with specific transcriptional characteristics and highlights the added value of distinguishing BP dual from single subtypes as evidenced by distinct treatment response rates.
Keywords: BluePrint; Breast cancer; Genomic testing; Molecular subtypes; Single and dual subtypes.
© 2022. The Author(s).
Conflict of interest statement
All authors (MMK, AE, AB, JCH, RB, DW, ACM, WMA, LM, and AMG) are non-commercial employees of Agendia, the commercial entity that markets the 80-gene signature as BluePrint. AMG is named inventor on the patent for the 80-gene signature used in this study. No writing assistance was utilized in the production of this manuscript.
Figures



References
-
- Press O, Guzman R, Cervantes M, et al. Characterization of HER2 status by fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC) New York: Springer; 2014. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

