Ectopic expression of WRINKLED1 in rice improves lipid biosynthesis but retards plant growth and development
- PMID: 35984829
- PMCID: PMC9390937
- DOI: 10.1371/journal.pone.0267684
Ectopic expression of WRINKLED1 in rice improves lipid biosynthesis but retards plant growth and development
Abstract
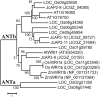
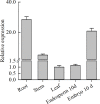
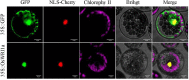
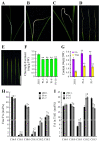
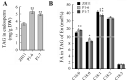
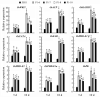
WRINKLED1 (WRI1) is a transcription factor which is key to the regulation of seed oil biosynthesis in Arabidopsis. In the study, we identified two WRI1 genes in rice, named OsWRI1a and OsWRI1b, which share over 98% nucleotide similarity and are expressed only at very low levels in leaves and endosperms. The subcellular localization of Arabidopsis protoplasts showed that OsWRI1a encoded a nuclear localized protein. Overexpression of OsWRI1a under the control of the CaMV 35S promoter severely retarded plant growth and development in rice. Expressing the OsWRI1a gene under the control of the P1 promoter of Brittle2 (highly expressed in endosperm but low in leaves and roots) increased the oil content of both leaves and endosperms and upregulated the expression of several genes related to late glycolysis and fatty acid biosynthesis. However, the growth and development of the transgenic plants were also affected, with phenotypes including smaller plant size, later heading time, and fewer and lighter grains. The laminae (especially those of flag leaves) did not turn green and could not unroll normally. Thus, ectopic expression of OsWRI1a in rice enhances oil biosynthesis, but also leads to abnormal plant growth and development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Bioch. 2002; 40(2):151–160. doi: 10.1016/S0981-9428(01)01350-X - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

