Mitochondrial mRNA localization is governed by translation kinetics and spatial transport
- PMID: 35984860
- PMCID: PMC9432724
- DOI: 10.1371/journal.pcbi.1010413
Mitochondrial mRNA localization is governed by translation kinetics and spatial transport
Abstract
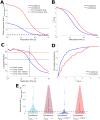
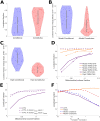
For many nuclear-encoded mitochondrial genes, mRNA localizes to the mitochondrial surface co-translationally, aided by the association of a mitochondrial targeting sequence (MTS) on the nascent peptide with the mitochondrial import complex. For a subset of these co-translationally localized mRNAs, their localization is dependent on the metabolic state of the cell, while others are constitutively localized. To explore the differences between these two mRNA types we developed a stochastic, quantitative model for MTS-mediated mRNA localization to mitochondria in yeast cells. This model includes translation, applying gene-specific kinetics derived from experimental data; and diffusion in the cytosol. Even though both mRNA types are co-translationally localized we found that the steady state number, or density, of ribosomes along an mRNA was insufficient to differentiate the two mRNA types. Instead, conditionally-localized mRNAs have faster translation kinetics which modulate localization in combination with changes to diffusive search kinetics across metabolic states. Our model also suggests that the MTS requires a maturation time to become competent to bind mitochondria. Our work indicates that yeast cells can regulate mRNA localization to mitochondria by controlling mitochondrial volume fraction (influencing diffusive search times) and gene translation kinetics (adjusting mRNA binding competence) without the need for mRNA-specific binding proteins. These results shed light on both global and gene-specific mechanisms that enable cells to alter mRNA localization in response to changing metabolic conditions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae.RNA. 2011 Aug;17(8):1551-65. doi: 10.1261/rna.2621111. Epub 2011 Jun 24. RNA. 2011. PMID: 21705432 Free PMC article.
-
Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization.PLoS One. 2008 Jun 4;3(6):e2293. doi: 10.1371/journal.pone.0002293. PLoS One. 2008. PMID: 18523582 Free PMC article.
-
eIF5A controls mitoprotein import by relieving ribosome stalling at TIM50 translocase mRNA.J Cell Biol. 2024 Dec 2;223(12):e202404094. doi: 10.1083/jcb.202404094. Epub 2024 Nov 7. J Cell Biol. 2024. PMID: 39509053 Free PMC article.
-
Here, there, everywhere. mRNA localization in budding yeast.RNA Biol. 2014;11(8):1031-9. doi: 10.4161/rna.29945. Epub 2014 Oct 31. RNA Biol. 2014. PMID: 25482891 Free PMC article. Review.
-
RNA localization in yeast: moving towards a mechanism.Biol Cell. 2005 Jan;97(1):75-86. doi: 10.1042/BC20040066. Biol Cell. 2005. PMID: 15601259 Review.
Cited by
-
Localization of RNAs to the mitochondria-mechanisms and functions.RNA. 2024 May 16;30(6):597-608. doi: 10.1261/rna.079999.124. RNA. 2024. PMID: 38448244 Free PMC article. Review.
-
Mitochondrial protein heterogeneity stems from the stochastic nature of co-translational protein targeting in cell senescence.Nat Commun. 2024 Sep 27;15(1):8274. doi: 10.1038/s41467-024-52183-y. Nat Commun. 2024. PMID: 39333462 Free PMC article.
-
Effects of axon branching and asymmetry between the branches on transport, mean age, and age density distributions of mitochondria in neurons: A computational study.Int J Numer Method Biomed Eng. 2022 Nov;38(11):e3648. doi: 10.1002/cnm.3648. Epub 2022 Oct 8. Int J Numer Method Biomed Eng. 2022. PMID: 36125402 Free PMC article.
-
Molecular machineries and pathways of mitochondrial protein transport.Nat Rev Mol Cell Biol. 2025 Jul 3. doi: 10.1038/s41580-025-00865-w. Online ahead of print. Nat Rev Mol Cell Biol. 2025. PMID: 40610778 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

