Intermediates in SARS-CoV-2 spike-mediated cell entry
- PMID: 35984891
- PMCID: PMC9390989
- DOI: 10.1126/sciadv.abo3153
Intermediates in SARS-CoV-2 spike-mediated cell entry
Abstract
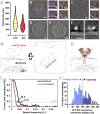
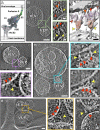
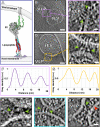
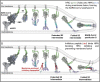
SARS-CoV-2 cell entry is completed after viral spike (S) protein-mediated membrane fusion between viral and host cell membranes. Stable prefusion and postfusion S structures have been resolved by cryo-electron microscopy and cryo-electron tomography, but the refolding intermediates on the fusion pathway are transient and have not been examined. We used an antiviral lipopeptide entry inhibitor to arrest S protein refolding and thereby capture intermediates as S proteins interact with hACE2 and fusion-activating proteases on cell-derived target membranes. Cryo-electron tomography imaged both extended and partially folded intermediate states of S2, as well as a novel late-stage conformation on the pathway to membrane fusion. The intermediates now identified in this dynamic S protein-directed fusion provide mechanistic insights that may guide the design of CoV entry inhibitors.
Figures


References
-
- Outlaw V. K., Bovier F. T., Mears M. C., Cajimat M. N., Zhu Y., Lin M. J., Addetia A., Lieberman N. A. P., Peddu V., Xie X., Shi P. Y., Greninger A. L., Gellman S. H., Bente D. A., Moscona A., Porotto M., Inhibition of coronavirus entry in vitro and ex vivo by a lipid-conjugated peptide derived from the SARS-CoV-2 spike glycoprotein HRC domain. MBio 11, e01935-20 (2020). - PMC - PubMed
-
- de Vries R. D., Schmitz K. S., Bovier F. T., Predella C., Khao J., Noack D., Haagmans B. L., Herfst S., Stearns K. N., Drew-Bear J., Biswas S., Rockx B., McGill G., Dorrello N. V., Gellman S. H., Alabi C. A., de Swart R. L., Moscona A., Porotto M., Intranasal fusion inhibitory lipopeptide prevents direct-contact SARS-CoV-2 transmission in ferrets. Science 371, 1379–1382 (2021). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

