PDIA3 epitope-driven immune autoreactivity contributes to hepatic damage in type 2 diabetes
- PMID: 35984892
- PMCID: PMC9762167
- DOI: 10.1126/sciimmunol.abl3795
PDIA3 epitope-driven immune autoreactivity contributes to hepatic damage in type 2 diabetes
Abstract
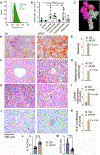
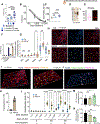
A diet rich in saturated fat and carbohydrates causes low-grade chronic inflammation in several organs, including the liver, ultimately driving nonalcoholic steatohepatitis. In this setting, environment-driven lipotoxicity and glucotoxicity induce liver damage, which promotes dendritic cell activation and generates a major histocompatibility complex class II (MHC-II) immunopeptidome enriched with peptides derived from proteins involved in cellular metabolism, oxidative phosphorylation, and the stress responses. Here, we demonstrated that lipotoxicity and glucotoxicity, as driven by a high-fat and high-fructose (HFHF) diet, promoted MHC-II presentation of nested T and B cell epitopes from protein disulfide isomerase family A member 3 (PDIA3), which is involved in immunogenic cell death. Increased MHC-II presentation of PDIA3 peptides was associated with antigen-specific proliferation of hepatic CD4+ immune infiltrates and isotype switch of anti-PDIA3 antibodies from IgM to IgG3, indicative of cellular and humoral PDIA3 autoreactivity. Passive transfer of PDIA3-specific T cells or PDIA3-specific antibodies also exacerbated hepatocyte death, as determined by increased hepatic transaminases detected in the sera of mice subjected to an HFHF but not control diet. Increased humoral responses to PDIA3 were also observed in patients with chronic inflammatory liver conditions, including autoimmune hepatitis, primary biliary cholangitis, and type 2 diabetes. Together, our data indicated that metabolic insults caused by an HFHF diet elicited liver damage and promoted pathogenic immune autoreactivity driven by T and B cell PDIA3 epitopes.
Conflict of interest statement
Figures





Comment in
-
Obesity-associated autoreactivity.Nat Rev Immunol. 2022 Oct;22(10):594-595. doi: 10.1038/s41577-022-00783-4. Nat Rev Immunol. 2022. PMID: 36071286 No abstract available.
-
PDIA3 as a potential bridge between immunogenic cell death and autoreactivity.Oncoimmunology. 2022 Sep 30;11(1):2130558. doi: 10.1080/2162402X.2022.2130558. eCollection 2022. Oncoimmunology. 2022. PMID: 36211809 Free PMC article. No abstract available.
References
-
- Hotamisligil GS, Shargill NS, Spiegelman BM, Adipose expression of tumor necrosis factor-alpha: Direct role in obesity-linked insulin resistance. Science 259, 87–91 (1993). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

