DHFR Mutants Modulate Their Synchronized Dynamics with the Substrate by Shifting Hydrogen Bond Occupancies
- PMID: 35984987
- PMCID: PMC9795552
- DOI: 10.1021/acs.jcim.2c00507
DHFR Mutants Modulate Their Synchronized Dynamics with the Substrate by Shifting Hydrogen Bond Occupancies
Abstract
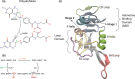
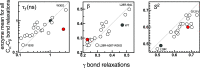
Antibiotic resistance is a global health problem in which mutations occurring in functional proteins render drugs ineffective. The working mechanisms of the arising mutants are seldom apparent; a methodology to decipher these mechanisms systematically would render devising therapies to control the arising mutational pathways possible. Here we utilize Cα-Cβ bond vector relaxations obtained from moderate length MD trajectories to determine conduits for functionality of the resistance conferring mutants of Escherichia coli dihydrofolate reductase. We find that the whole enzyme is synchronized to the motions of the substrate, irrespective of the mutation introducing gain-of-function or loss-of function. The total coordination of the motions suggests changes in the hydrogen bond dynamics with respect to the wild type as a possible route to determine and classify the mode-of-action of individual mutants. As a result, nine trimethoprim-resistant point mutations arising frequently in evolution experiments are categorized. One group of mutants that display the largest occurrence (L28R, W30G) work directly by modifying the dihydrofolate binding region. Conversely, W30R works indirectly by the formation of the E139-R30 salt bridge which releases energy resulting from tight binding by distorting the binding cavity. A third group (D27E, F153S, I94L) arising as single, resistance invoking mutants in evolution experiment trajectories allosterically and dynamically affects a hydrogen bonding motif formed at residues 59-69-71 which in turn modifies the binding site dynamics. The final group (I5F, A26T, R98P) consists of those mutants that have properties most similar to the wild type; these only appear after one of the other mutants is fixed on the protein structure and therefore display clear epistasis. Thus, we show that the binding event is governed by the entire enzyme dynamics while the binding site residues play gating roles. The adjustments made in the total enzyme in response to point mutations are what make quantifying and pinpointing their effect a hard problem. Here, we show that hydrogen bond dynamics recorded on sub-μs time scales provide the necessary fingerprints to decipher the various mechanisms at play.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

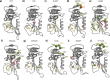
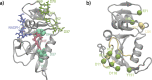
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

