Evaluation of indirect sequence-specific magneto-extraction-aided LAMP for fluorescence and electrochemical SARS-CoV-2 nucleic acid detection
- PMID: 35985192
- PMCID: PMC9373715
- DOI: 10.1016/j.talanta.2022.123809
Evaluation of indirect sequence-specific magneto-extraction-aided LAMP for fluorescence and electrochemical SARS-CoV-2 nucleic acid detection
Abstract
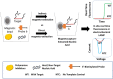
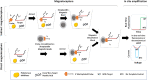
Nucleic acid amplification tests (NAATs) such as quantitative real-time reverse transcriptase PCR (qRT-PCR) or isothermal NAATs (iNAATs) such as loop-mediated isothermal amplification (LAMP) require pure nucleic acid free of any polymerase inhibitors as its substrate. This in turn, warrants the use of spin-column mediated extraction with centralized high-speed centrifuges. Additionally, the utilization of centralized real-time fluorescence readout and TaqMan-like molecular probes in qRT-PCR and real-time LAMP add cost and restrict their deployment. To circumvent these disadvantages, we report a novel sample-to-answer workflow comprising an indirect sequence-specific magneto-extraction (also referred to as magnetocapture, magneto-preconcentration, or magneto-enrichment) for detecting SARS-CoV-2 nucleic acid. It was followed by in situ fluorescence or electrochemical LAMP. After in silico validation of the approach's sequence selectivity against SARS-CoV-2 variants of concern, the comparative performance of indirect and direct magnetocapture in detecting SARS-CoV-2 nucleic acid in the presence of excess host nucleic acid or serum was probed. After proven superior, the sensitivity of the indirect sequence-specific magnetocapture in conjunction with electrochemical LAMP was investigated. In each case, its sensitivity was assessed through the detection of clinically relevant 102 and 103 copies of target nucleic acid. Overall, a highly specific nucleic acid detection method was established that can be accommodated for either centralized real-time SYBR-based fluorescence LAMP or portable electrochemical LAMP.
Keywords: Electrochemical LAMP; Magnetic sequence-specific target enrichment; Quantitative real-time LAMP; SARS-CoV-2.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Dr. Souradyuti Ghosh reports financial support was provided by Mission on Nano Science and Technology - Department of Science and Technology-Government of India for Nanomission grant. Dr. Souradyuti Ghosh reports financial support was provided by Department of Biotechnology, India grant. Dr. Souradyuti Ghosh has patent #202111028722 pending to Bennett University, India. Dr. Souradyuti Ghosh has patent #202111037358 pending to Bennett University.
Figures




References
-
- Johns Hopkins Coronavirus Resource Center: Home, Johns Hopkins Coronavirus Resour. Cent. (n.d.). https://coronavirus.jhu.edu/(accessed January 24, 2022).
-
- Q. Diagnostics, FDA instructions on detectimg SARS-CoV-2 RNA with real-time RT-PCR, (n.d.). https://www.fda.gov/media/136231/download.
-
- RT-PCR: One step vs. two step | IDT, Integr. DNA Technol. (n.d.). https://sg.idtdna.com/pages/support/faqs/what-are-the-advantages-and-dis... (accessed December 4, 2021).
-
- Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Barreto Miranda I., Schnitzler P., Kräusslich H.-G., Knop M., Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc7075. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

