Emergence of a multidrug-resistant and virulent Streptococcus pneumoniae lineage mediates serotype replacement after PCV13: an international whole-genome sequencing study
- PMID: 35985351
- PMCID: PMC9519462
- DOI: 10.1016/S2666-5247(22)00158-6
Emergence of a multidrug-resistant and virulent Streptococcus pneumoniae lineage mediates serotype replacement after PCV13: an international whole-genome sequencing study
Abstract
Background: Serotype 24F is one of the emerging pneumococcal serotypes after the introduction of pneumococcal conjugate vaccine (PCV). We aimed to identify lineages driving the increase of serotype 24F in France and place these findings into a global context.
Methods: Whole-genome sequencing was performed on a collection of serotype 24F pneumococci from asymptomatic colonisation (n=229) and invasive disease (n=190) isolates among individuals younger than 18 years in France, from 2003 to 2018. To provide a global context, we included an additional collection of 24F isolates in the Global Pneumococcal Sequencing (GPS) project database for analysis. A Global Pneumococcal Sequence Cluster (GPSC) and a clonal complex (CC) were assigned to each genome. Phylogenetic, evolutionary, and spatiotemporal analysis were conducted using the same 24F collection and supplemented with a global collection of genomes belonging to the lineage of interest from the GPS project database (n=25 590).
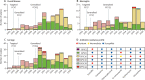
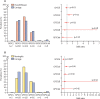
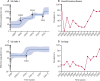
Findings: Serotype 24F was identified in numerous countries mainly due to the clonal spread of three lineages: GPSC10 (CC230), GPSC16 (CC156), and GPSC206 (CC7701). GPSC10 was the only multidrug-resistant lineage. GPSC10 drove the increase in 24F in France and had high invasive disease potential. The international dataset of GPSC10 (n=888) revealed that this lineage expressed 16 other serotypes, with only six included in 13-valent PCV (PCV13). All serotype 24F isolates were clustered in a single clade within the GPSC10 phylogeny and long-range transmissions were detected from Europe to other continents. Spatiotemporal analysis showed GPSC10-24F took 3-5 years to spread across France and a rapid change of serotype composition from PCV13 serotype 19A to 24F during the introduction of PCV13 was observed in neighbouring country Spain.
Interpretation: Our work reveals that GPSC10 alone is a challenge for serotype-based vaccine strategy. More systematic investigation to identify lineages like GPSC10 will better inform and improve next-generation preventive strategies against pneumococcal diseases.
Funding: Bill & Melinda Gates Foundation, Wellcome Sanger Institute, and the US Centers for Disease Control and Prevention.
Copyright © 2022 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests AvG reports grants from the US Centers for Disease Control and Prevention and Sanofi; travel fees from Pfizer, Sanofi, and Merck; and is the chairperson of South Africa National Advisory Group on Immunisation, outside of the submitted work. AJP reports grants from Gavi, WHO, and AstraZeneca, and is a member of the Joint Committee on Vaccination and Immunisation and WHO Strategic Advisory Group of Experts on Immunization, outside of the submitted work. IN reports grants from BMGF and travel fees from the International Symposium on Pneumococci and Pneumococcal Diseases, outside of the submitted work. JM is an employee of Pharma Vaccines, outside of the submitted work. RD reports grants from Pfizer, MSD, and MedImmune–AstraZeneca, consulting and speaker fees from Pfizer and MSD, outside of the submitted work. SDB reports personal fees from Pfizer and Merck, outside the submitted work. CMA reports grants from Pfizer, outside of the submitted work. EV report grants from Santé Publique France, Pfizer, and MSD, outside of the submitted work. All other authors declare no competing interests.
Figures

References
-
- Ouldali N, Levy C, Varon E, et al. Incidence of paediatric pneumococcal meningitis and emergence of new serotypes: a time-series analysis of a 16-year French national survey. Lancet Infect Dis. 2018;18:983–991. - PubMed
-
- Ouldali N, Varon E, Levy C, et al. Invasive pneumococcal disease incidence in children and adults in France during the pneumococcal conjugate vaccine era: an interrupted time-series analysis of data from a 17-year national prospective surveillance study. Lancet Infect Dis. 2021;21:137–147. - PubMed
-
- Pai R, Moore MR, Pilishvili T, Gertz RE, Whitney CG, Beall B. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J Infect Dis. 2005;192:1988–1995. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

